Figure 1.
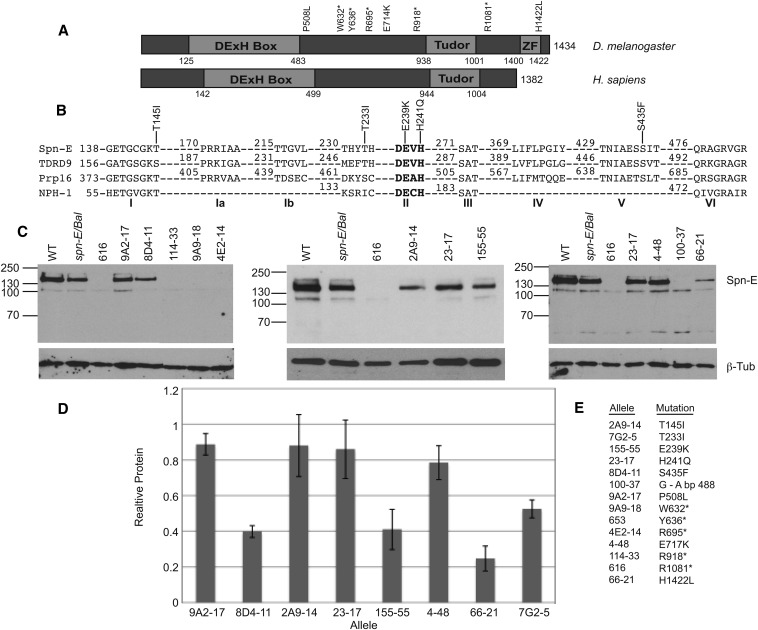
Eight of the fourteen spn-E alleles express detectable protein and have point mutations in the SPN-E coding region. (A) Domain structure of Drosophila SPN-E and its human homolog, TDRD9. SPN-E contains a highly conserved DExH box and a Tudor domain as well as a Zinc finger, whereas TDRD9 only has a DExH box and Tudor domain. The position of the two mutations outside of the conserved domains, the five mutations that do not produce detectable protein, and the Zinc finger are shown. (B) The amino acid sequence of the SPN-E DExH box domain compared with its human homolog TDRD9, yeast splicing factor Prp16, and vaccinia virus protein NPH-I. The positions of the five mutations identified in the SPN-E DExH box domain are shown. Amino acid numbering is according to Ensemble Genome Browser release 73. (C) SPN-E protein expression in mutant ovary extracts as measured by Western blotting. Protein was isolated from hemizygous ovaries of the genotype spn-Emutant/spn-E∆125. Eight alleles express detectable protein of the correct size for SPN-E. Four alleles do not express detectable protein. Spn-E/Bal = spn-E∆125/Balancer chromosome. Line 7G2-5 is not shown. Several extraneous bands are found on the Western blots shown above. We did not detect these bands when we used a second antibody developed in the laboratory of Dr. Toshie Kai (Patil and Kai 2010; data not shown); therefore, we think that the extra bands are most likely nonspecific bands recognized by our SPN-E antibody. (D) SPN-E protein levels in the various mutant ovaries relative to spn-E∆125/Balancer. Error bars represent SD of at least 2 separate protein isolates. SPN-E protein levels were normalized to beta-tubulin. (E) A listing of each spn-E allele name along with its corresponding mutation.