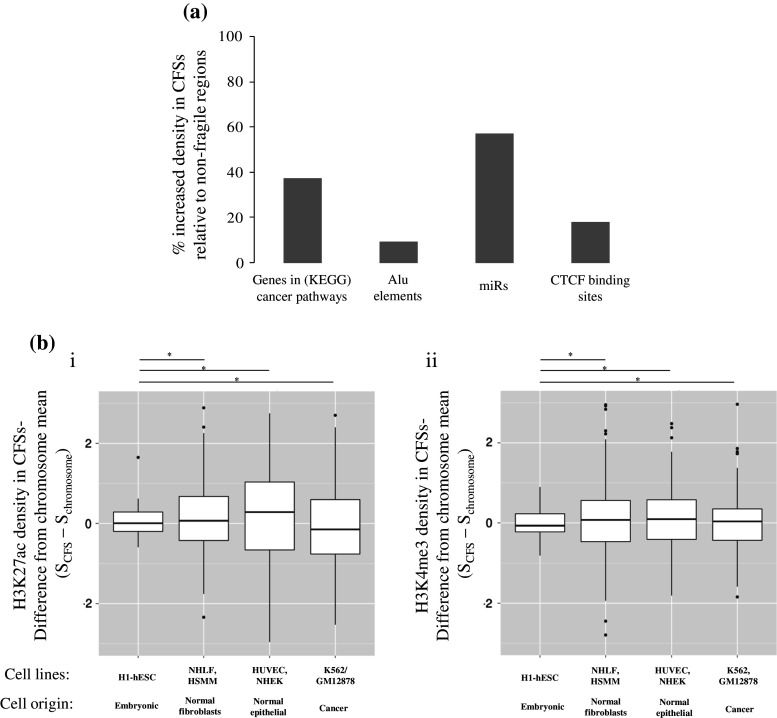
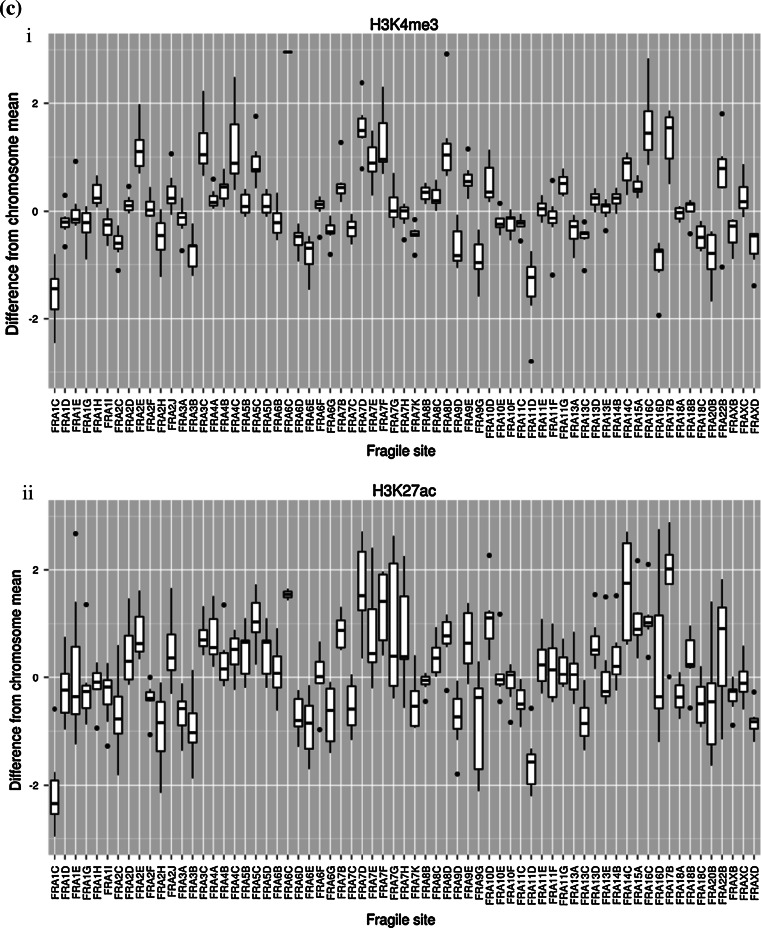
Fig. 2.
Frequency of cancer-related genes, repetitive elements, miRs, binding elements, and histone marks in CFSs. a CFSs exhibit a higher density of cancer-related genes (obtained from Kyoto Encyclopedia of Genes and Genomes), Alu repetitive elements [19], miRs, and the CTCF binding element relative to non-fragile regions. b CFSs exhibit a differential density of the histone marks (i) Histone 3 lysine 27 acetylation (H3K27ac) and (ii) Histone 3 lysine 4 trimethylation (H3K4me3), relative to non-fragile regions that is cell type origin-dependent (data concerning histone modifications derived from ChIP-seq experiments belonging to the ENCODE project were downloaded from the UCSC server (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegMarkH3k4me3/, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegMarkH3k27ac/). Specifically, we obtained bigWig files for H3k4me3 and H3k27ac modifications in the GM12878, H1-hesc, HSMM, HUVEC, K562, NHEK, and NHLF cell types. Information concerning regions of interest was extracted with the bigWigSummary utility, also available from the UCSC server. Specifically, the average signal was calculated for every chromosome in every cell line by repeatedly invoking: bigWigSummary-type = mean “bigwigfile” chrN start end. Similarly, the average signal was calculated for cytogenetically and molecularly mapped fragile sites. For every defined fragile site, the mean histone modification signal of the corresponding chromosome was subtracted from the mean signal of the fragile region. Signal difference from mean for site i = mean(FSi) − mean(chromosomeFSi). The chromosome means varied within each cell type (data not shown), as did the histone modifications between cell types) (S histone signal). c Frequency of histone marks per CFSs. Each CFS exhibits a differential density of the histone marks. (i) Histone 3 lysine 4 trimethylation (H3K4me3) and (ii) Histone 3 lysine 27 acetylation (H3K27ac), relative to non-fragile regions averaged over all cell types presented in Fig. 2 (using the data generated for Fig. 2b, we also plotted a boxplot for individual cytogenetically defined CFSs in all 7 cell lines mentioned above with respect to H3K4me3 and H3K27ac. Significant heterogeneity between CFSs can be observed)