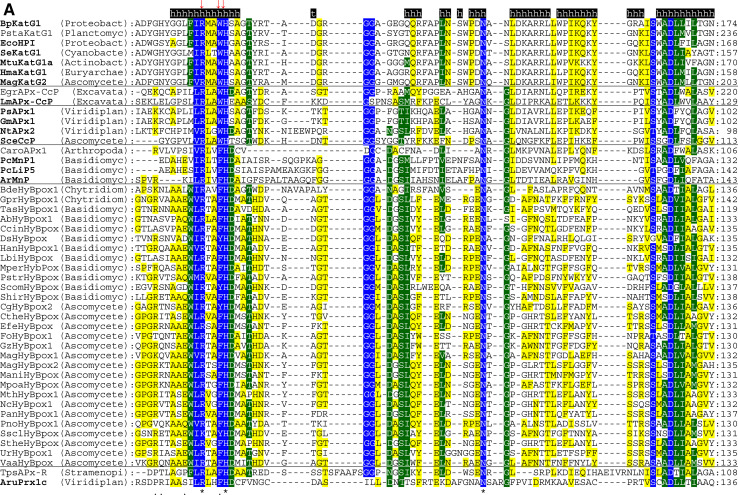
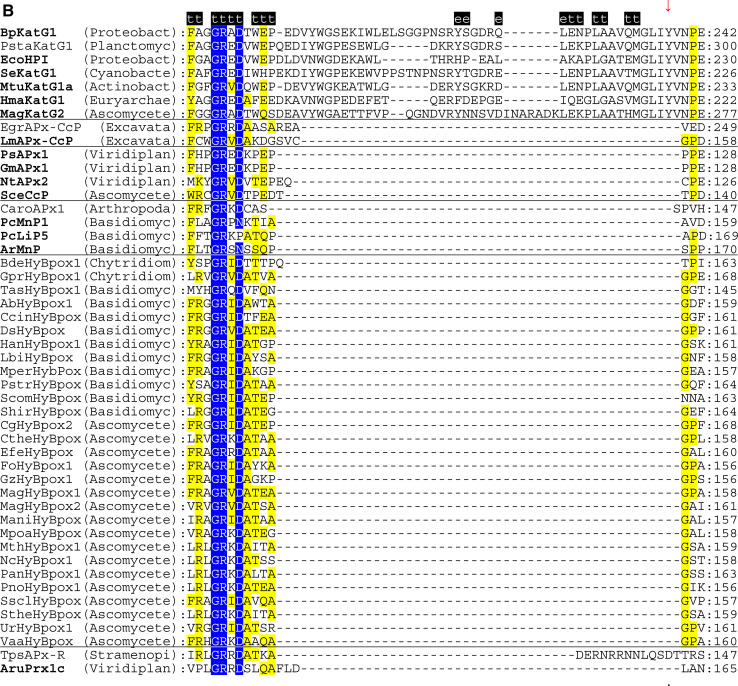
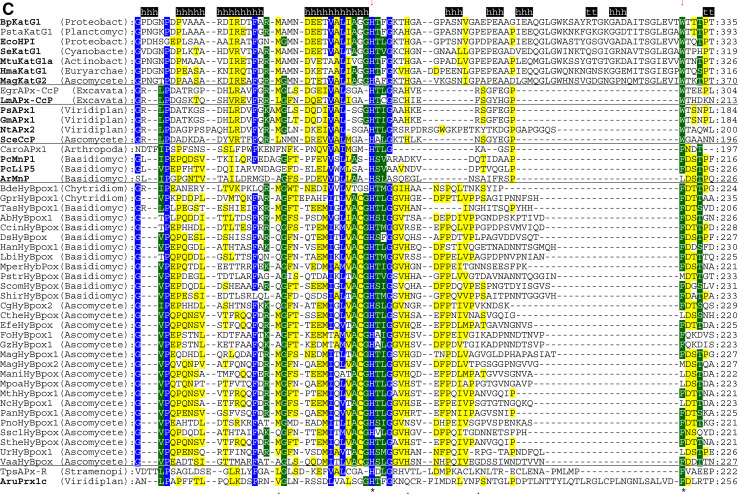
Fig. 5.
Selected parts of structural multiple sequence alignment of 48 members of the peroxidase–catalase superfamily. This alignment demonstrates both high conservation of the active site residues as well as some variability. a Region including residues at the distal heme side, b region of the large loop, c region including residues at the proximal heme side. Secondary structural elements taken from the 3D structure of KatG from Burkholderia pseudomallei (BpKatG, PDB code 1MWV) are depicted (h helix, e strand, t turn). Essential residues involved in catalysis are labeled as “*” and those residues that were involved in catalysis but later during the evolution mutated as “.” residues discussed in the text are labeled with arrows. Sequences with known 3D structures are in bold. Parameters for the alignment are described in the Sect. “Materials and methods”. Abbreviations of peroxidase names are taken from PeroxiBase