Figure 5.
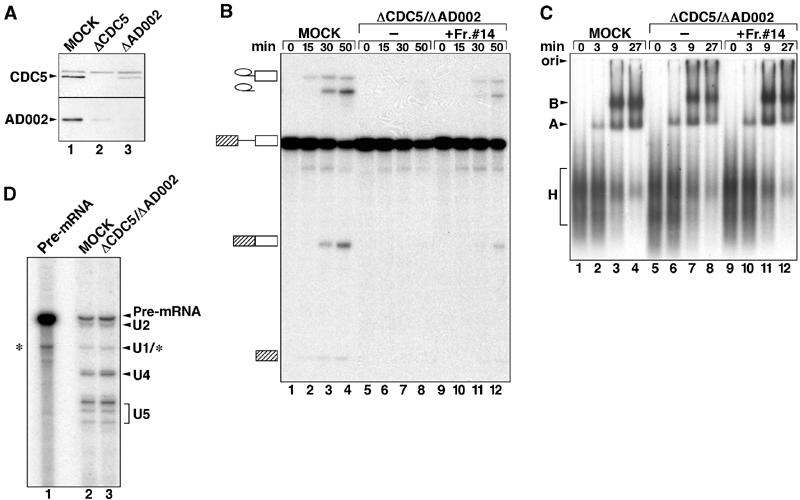
Spliceosomal B complex forms in the absence of the CDC5/PRP19 complex. (A) Nuclear extracts are efficiently immunodepleted of CDC5 and AD002. Western blot of mock-depleted (lane 1), CDC5-depleted (lane 2) or AD002-depleted (lane 3) extract probed with anti-CDC5 (upper panel) or anti-AD002 (lower panel) antibodies. (B) Time course of splicing of [32P] MINX pre-mRNA in mock-depleted extract (MOCK, lanes 1–4) or extract depleted with CDC5 and AD002 (ΔCDC5/ΔAD002, lanes 5–8), or CDC5/AD002-depleted extract complemented with purified CDC5/PRP19 complex (fraction no. 14, from the gradient in Figure 4) (lanes 9–12). RNA was analyzed by denaturing PAGE and visualized by autoradiography. The pre-mRNA, and splicing intermediates and products are indicated on the left. (C) The CDC5/PRP19 complex is not required for B-complex formation. Spliceosome assembly in mock-depleted extract, CDC5/AD002-depleted extract, or CDC5/AD002-depleted extract complemented with the CDC5/PRP19 complex (lanes marked as in (B)) was analyzed at the indicated times by native gel electrophoresis and visualized by autoradiography. The position of the H, A, and B complexes is indicated on the left. (D) Spliceosomes formed in CDC5/AD002-depleted extract contain U4 snRNA. Spliceosomes were immunoprecipitated from mock- (lane 2) or CDC5/AD002-depleted extract (lane 3) with anti-61K antibodies. RNA was isolated, end labeled with 32P-pCp, fractionated by denaturing PAGE, and visualized by autoradiography. The identities of the RNAs are indicated on the right. Internally radiolabeled MINX pre-mRNA (lane 1) was analyzed in parallel. Note that the band migrating at the position of U1 may also contain a pre-mRNA degradation product (*).
