Figure 5.
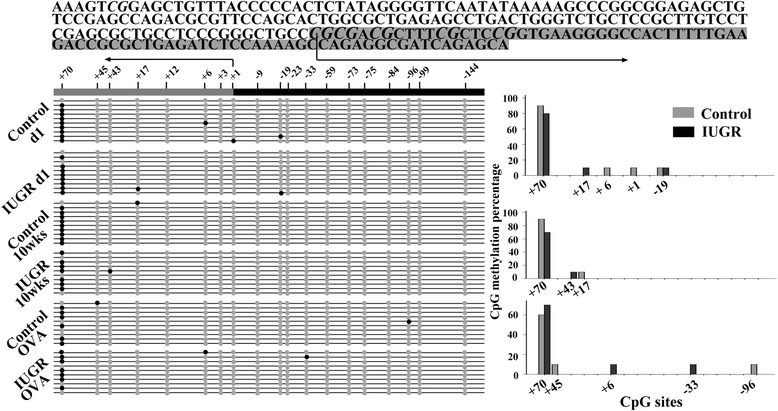
Methylation profiles of the ET-1 core promoter in lung tissue from IUGR and control rats are shown. The proximal promoter sequence of ET-1 (−149 to +80) is shown. Methylated CpG sites are indicated by solid black circles and unmethylated CpGs are depicted by grey circles. Arrows represent transcription start sites. Each line of circles represents independent PCR clones (n = 10). The bar graph represents CpG methylation percentage at each site, there was no significant difference between the Control and IUGR animals. Only the +70 site was hypermethylated, whereas sites +45, +43, +17, +6, +1, −19, −33, and −96 were hypomethylated, and the sites +12, +3, −9, −23, −59, −73, −75, −99, and −144 were unmethylated. There were no significantly statistical differences between Control and IUGR rats (P >0.05) at any of CpG sites.
