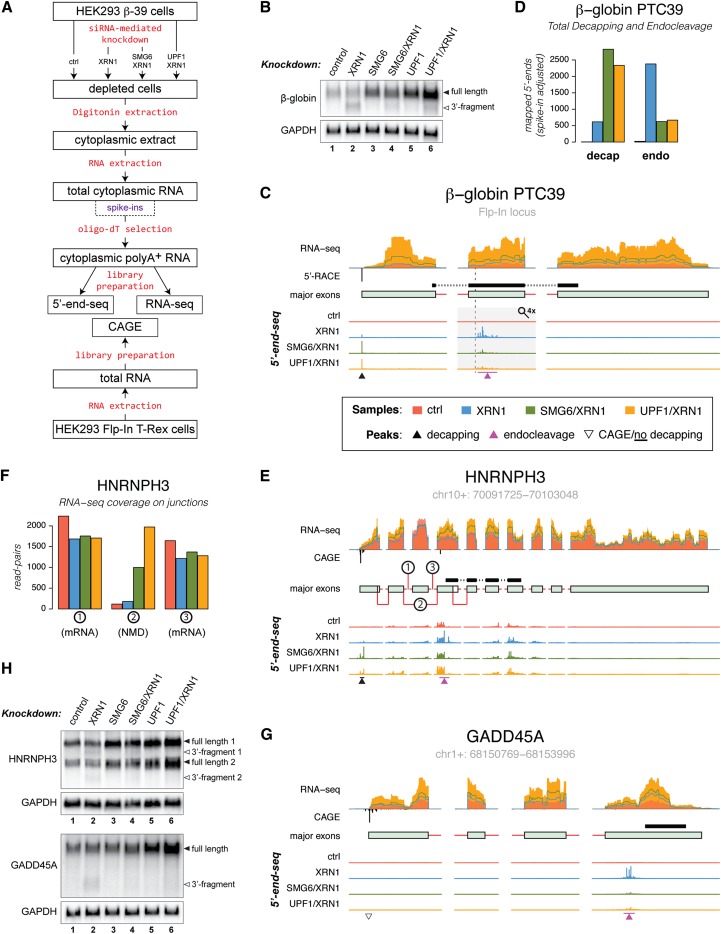
Figure 1.
NMD-specific endonucleolytic cleavage sites are revealed by 5′ end-seq. (A) Schematic outline of the sample preparation steps for the massive parallel sequencing procedures used in this study (see the text and Supplemental Fig. S1 for details). (B) Northern blotting analysis of total RNA isolated from HEK293-β-39 cells depleted for the indicated factors. The Northern membrane was hybridized with a probe directed against the region shown in C. GAPDH levels were detected as an internal loading standard. (C) Overview of the stably integrated β-39 gene illustrating the sequencing data used in the analyses. From top to bottom, the tracks display (1) RNA-seq coverage over the major exons as well as leading and trailing intronic sequences determined from control, XRN1-depleted, SMG6/XRN1-depleted, and UPF1/XRN1-depleted samples (in all figures, data from the control, XRN1-depleted, SMG6/XRN1-depleted, and UPF1/XRN1-depleted samples are depicted in red, blue, green, and orange, respectively). (2) 5′-RACE-mapped 5′ end of the transcript (black; below RNA-seq axis). For all endogenous genes, 5′ ends were determined by CAGE. (3) Schematic representation of the major exons expressed from the gene (exons and intronic sequences are represented as light-green boxes and red lines, respectively; see the Materials and Methods for details). The position of the probe used for Northern blotting is shown as a black bar above the transcript model. The PTC is indicated by a vertical dashed line. (4) 5′ end-seq-determined 5′ end signals displayed in individual tracks but on the same scale for control, XRN1-depleted, SMG6/XRN1-depleted, and UPF1/XRN1-depleted samples. To allow simultaneous visualization of decapping and endocleavage peaks (indicated by black and purple arrowheads, respectively), the 5′ end-seq tracks were scaled up by a factor of 4 in the region covering exon 2 of the β-39 RNA (indicated by a gray rectangle and a magnifying glass). (D) Histogram showing total signal in the decapping peaks (left) and NMD-specific endocleavage sites (right). (E,G) As in C, but for the HNRNPH3 and GADD45A loci. The open triangle in G indicates a transcript 5′ end determined by CAGE for which no decapping signal was detected. (F) Histogram showing the RNA-seq coverage over the exon–exon junctions indicated in E. (H) As in B, but the membranes were hybridized with probes directed against HNRNPH3 (top panel) and GADD45A (bottom panel) RNA species (see E and G for positions of probes). See also Supplemental Figure S2.