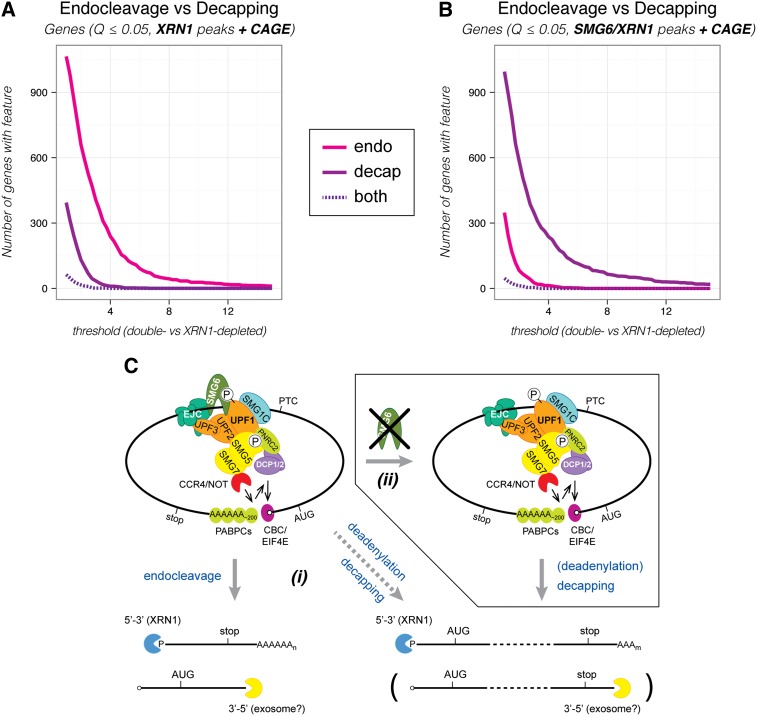
Figure 3.
NMD-specific endocleavage dominates over decapping in HEK293 cells. (A) The number of genes producing one or more transcripts with one or more endonucleolytic cleavage sites (endo [magenta]), decapping sites (decap [purple]), or both (dashed, dark purple) as a function of the threshold used to define the given event (see the text for details). The XRN1 depletion sample was compared with the control sample to identify potential sites in transcripts with CAGE information in order to allow a “fair” comparison between endocleavage and decapping events (XRN1 peaks + CAGE). We note that the dominance of endocleavage over decapping was robust over a range of applied criteria for the initial peak detection. The NMD-specific endocleavage and decapping events were determined without taking differential expression determined by RNA-seq data into account. (B) As in A, but with potential sites initially identified in the SMG6/XRN1 depletion sample (SMG6/XRN1 peaks + CAGE). Analyses corresponding to A and B, but done for transcripts instead of genes is shown in Supplemental Figure S5, A and B. See Supplemental Figure S4 for details about peak identification. (C) Model for NMD in humans. The translation machinery stalls (not shown) when it encounters a PTC. The termination codon is marked as premature by a protein complex that includes UPF1, UPF2, and UPF3. Phosphorylation of UPF1 by SMG1C commits the RNA to NMD. (Left top panel) Subsequently, a set of proteins (SMG5/SMG7, PNRC2, and/or SMG6) that deprotects and prepares the RNA for exonucleolytic degradation is recruited to phosphorylated UPF1. SMG6 catalyzes a local PTC-proximal endocleavage (left bottom panel), whereas SMG5/SMG7 interacts with the CCR4/NOT deadenylation complex that catalyzes polyA tail shortening, which in turn stimulates decapping (right bottom panel). Furthermore, UPF1 can interact with the decapping complex (DCP1/2) and stimulate decapping either directly or indirectly via binding to PNRC2. (i) We suggest that SMG6-mediated endocleavage is the first and fastest response, whereas decapping is kinetically less favored. (ii) However, decapping can partially substitute for endocleavage if SMG6 function is somehow hindered (as seen upon depletion of SMG6). See the Discussion for further details.