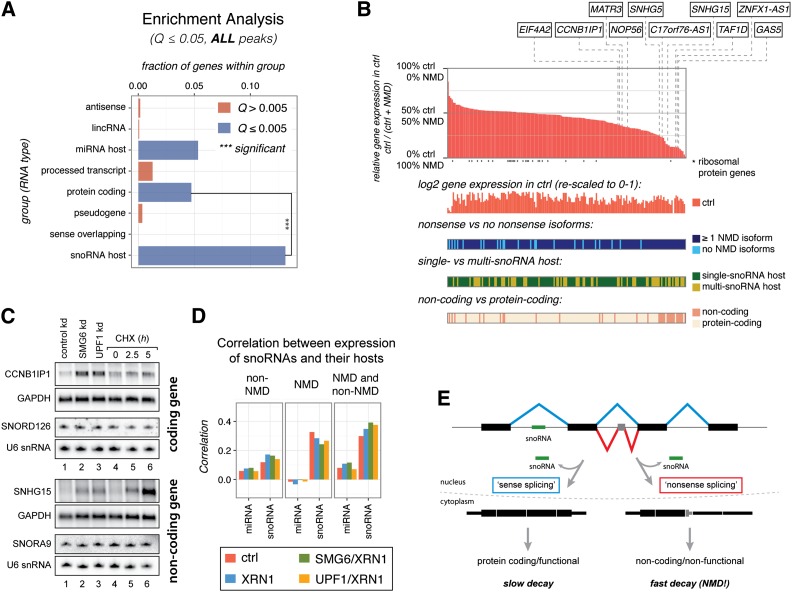
Figure 5.
snoRNA host genes are enriched among NMD substrates. (A) Enrichment analysis for the indicated RNA biotypes in the “stringent” NMD target gene set. Bar sizes indicate the fraction of genes in the given category that is NMD substrates. Blue bars indicate that there is a significant enrichment (Fisher one-sided test, Benjamini-Hochberg-corrected Q < 0.005). Three asterisks indicate that snoRNA host genes are significantly enriched compared with protein-coding genes (Fisher two-sided test, P = 2.1 × 10−10). A similar analysis based on “relaxed” criteria is presented in Supplemental Figure S5E. (B) Summarized features of human snoRNA host genes. (Top panel) The main plot represents the relative gene expression levels of the 173 analyzed human snoRNA host genes compared between the control sample and the mean value obtained from the SMG6/XRN1 and UPF1/XRN1 samples (NMD). Individual verified genes are pointed out by dashed lines. Asterisks below the X-axis indicate genes encoding ribosomal proteins. Four additional plots below the main plot show, from top to bottom, (1) the log2-transformed absolute expression levels from the control sample (rescaled to a range of 0–1), (2) whether nonsense RNA isoforms are produced from the gene, (3) whether the gene is a single- or a multi-snoRNA host, and (4) whether the gene is annotated as protein-coding or noncoding. (C) Northern blotting analyses of total RNA isolated from the HEK293-β-39 cell line depleted for the indicated factors (lanes 1–3) or incubated for 0, 2.5, and 5 h with the translation inhibitor cycloheximide (CHX) (lanes 4–6). The membranes were hybridized with probes directed against RNA from the snoRNA host genes CCNB1IP1 (protein-coding) and SNHG15 (noncoding), respectively. GAPDH levels were detected as an internal loading standard. Separate Northern blots were probed for corresponding intron-encoded snoRNAs (SNORD126 and SNORA9) and U6 snRNA as an internal small RNA loading standard. Slowed decay under UPF1 depletion conditions is demonstrated for the SNHG15 transcript in Supplemental Figure S2G. (D) Correlation coefficients between the steady-state expression levels of small RNAs and their hosts. The left cluster in each panel shows the correlation between miRNA host transcript expression determined from the four RNA-seq libraries and the encoded miRNAs, and the right cluster shows the correlation between the snoRNA hosts and their encoded snoRNAs. The expression of small RNAs was correlated to a group of NMD-insensitive transcripts (left panel), NMD-sensitive transcripts (middle panel), and both groups together (right panel). (E) Model illustrating that a snoRNA host gene can produce both spliced stable and nonsense RNA, which in both cases leads to production of the encoded snoRNA (see the text for details). See also Supplemental Figure S6.