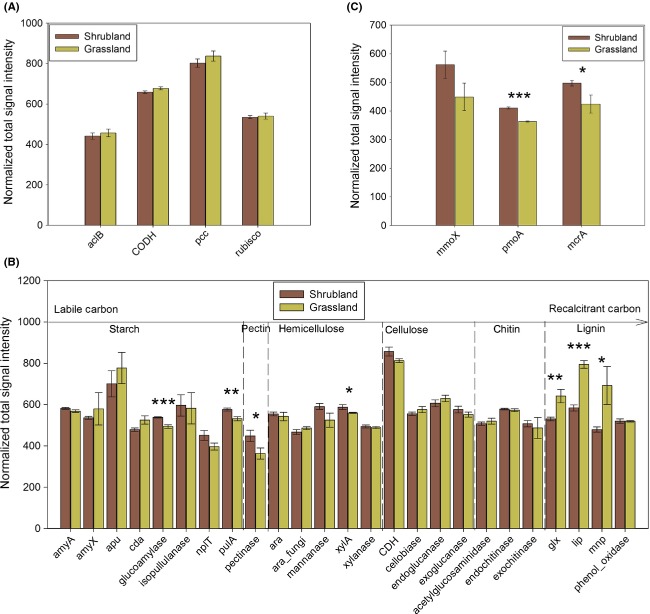
Figure 1.
The total abundance of (A) carbon fixation, (B) carbon degradation, and (C) methane cycling genes in the shrubland and grassland. Error bars represented standard error (n = 3). The differences between the shrubland and the grassland sites were examined by two-tailed paired t-tests. “*” P < 0.10, “**” P < 0.05, “***” P < 0.01. aclB, ATP-citrate lyase beta subunit; CODH, carbon monoxide dehydrogenase; pcc, propionyl-CoA carboxylase; rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase; mmoX, methane mono-oxygenase; pmoA, particulate methane mono-xygenase alpha subunit; mcrA, methyl-coenzyme M reductase alpha subunit; amyA, alpha-amylase; amyX, pullulanase; apu, amylopullulanase; cda, cytidine deaminase; nplT, neopullulanase; pulA, glycogen debranching enzyme; ara, alpha-N-arabinofuranosidase; xylA, xylose isomerase; CDH, cellobiose dehydrogenase; glx, glyoxalase; lip, triacylglycerol lipase; mnp, and manganese peroxidase.