Abstract
Arginine decarboxylase produces agmatine, and arginase and agmatinase are ureohydrolases that catalyze the production of ornithine and putrescine from arginine and agmatine, respectively, releasing urea. In the genome of the filamentous, heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120, ORF alr2310 putatively encodes an ureohydrolase. Cells of Anabaena supplemented with [14C]arginine took up and catabolized this amino acid generating a set of labeled amino acids that included ornithine, proline, and glutamate. In an alr2310 deletion mutant, an agmatine spot appeared and labeled glutamate increased with respect to the wild type, suggesting that Alr2310 is an agmatinase rather than an arginase. As determined in cell-free extracts, agmatinase activity could be detected in the wild type but not in the mutant. Thus, alr2310 is the Anabaena speB gene encoding agmatinase. The Δalr2310 mutant accumulated large amounts of cyanophycin granule polypeptide, lacked nitrogenase activity, and did not grow diazotrophically. Growth tests in solid media showed that agmatine is inhibitory for Anabaena, especially under diazotrophic conditions, suggesting that growth of the mutant is inhibited by non-metabolized agmatine. Measurements of incorporation of radioactivity from [14C]leucine into macromolecules showed, however, a limited inhibition of protein synthesis in the Δalr2310 mutant. Analysis of an Anabaena strain producing an Alr2310-GFP (green fluorescent protein) fusion showed expression in vegetative cells but much less in heterocysts, implying compartmentalization of the arginine decarboxylation pathway in the diazotrophic filaments of this heterocyst-forming cyanobacterium.
Keywords: Agmatinase, Anabaena, arginine catabolism, nitrogenase
Introduction
Cyanobacteria are oxygenic photosynthetic prokaryotes that constitute a phylogenetically coherent group of organisms (Giovannoni et al. 1988). However, they show very diverse morphologies, presenting both unicellular and multicellular forms (Rippka et al. 1979). Cyanobacteria such as those of the genera Anabaena and Nostoc grow as filaments of cells (trichomes) that, when incubated in the absence of a source of combined nitrogen, present two cell types: vegetative cells that fix CO2 performing oxygenic photosynthesis and heterocysts that carry out N2 fixation (Flores and Herrero 2010). Heterocysts differentiate from vegetative cells in a process that involves execution of a specific program of gene expression and intercellular transfer of regulators (Herrero et al. 2013). In the N2-fixing filament, heterocysts are spaced along the filament and provide the vegetative cells with fixed nitrogen; for this, heterocysts need to receive in turn photosynthate from the vegetative cells (Wolk et al. 1994). An intercellular exchange of glutamine for glutamate (Thomas et al. 1977; Martín-Figueroa et al. 2000) and transfer of β-aspartyl-arginine (Burnat et al. 2014) can move fixed nitrogen from heterocysts to vegetative cells. Conversely, alanine (Jüttner 1983; Pernil et al. 2010) and, mainly, sucrose (Jüttner 1983; Schilling and Ehrnsperger 1985; Curatti et al. 2002; López-Igual et al. 2010; Vargas et al. 2011) can transfer reduced carbon from vegetative cells to heterocysts.
Heterocysts conspicuously accumulate cyanophycin (multi-l-arginyl-poly [l-aspartic acid]), a nitrogen-rich reserve polymer (Lang et al. 1972; Sherman et al. 2000), which is seen as refractile granules that are located at the heterocyst poles (close to and inside the heterocyst “necks”). Although production of cyanophycin is not required for diazotrophic growth (Ziegler et al. 2001; Picossi et al. 2004), its conspicuous presence in the heterocysts suggests a possible role in diazotrophy, likely as dynamic nitrogen storage (Carr 1983; Haselkorn 2007). Cyanophycin is synthesized by cyanophycin synthetase and degraded by cyanophycinase, which releases β-aspartyl-arginine (Richter et al. 1999). In the diazotrophic filaments, both cyanophycin synthetase and cyanophycinase are present at high levels in the heterocysts (Gupta and Carr 1981a; Picossi et al. 2004). The β-aspartyl-arginine dipeptide is hydrolyzed to aspartate and arginine by an isoaspartyl dipeptidase (Hejazi et al. 2002), which in the model heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120 (hereafter Anabaena) has been found to be expressed preferentially in vegetative cells (Burnat et al. 2014). This implies that the dipeptide released by cyanophycinase in the heterocyst is transferred to vegetative cells serving as a nitrogen vehicle in the diazotrophic filament. The isoaspartyl dipeptidase products, arginine and aspartate, must be further catabolized to make their nitrogen atoms available for cellular metabolism.
Arginine serves as a source of nitrogen, carbon, and energy in different bacteria, and five possible catabolic pathways have been detected in cyanobacteria by bioinformatic analysis (Schriek et al. 2007). Of these, three (shown schematically in Fig.1) have been detected by physiological studies: the arginase pathway, the arginine deiminase pathway, and the arginine decarboxylase pathway (Quintero et al. 2000; Schriek et al. 2007; Weathers et al. 2010). In the arginase pathway, arginine is converted to ornithine releasing urea, and ornithine is further metabolized to glutamate (Cunin et al. 1986). Arginine deiminase produces citrulline and ammonium, with citrulline being further catabolized producing ornithine and carbamoyl phosphate (Cunin et al. 1986). In the arginine decarboxylase pathway, arginine is decarboxylated to agmatine (1-[4-aminobutyl]guanidine), which is then metabolized to putrescine (1,4-diaminobutane) releasing urea and further to 4-aminobutyrate and succinate (Cunin et al. 1986). In Aphanocapsa 6308 (currently known as Synechocystis sp. strain PCC 6308), it was reported that, besides the arginase pathway, which would provide only nitrogen for the cells, the arginine deiminase (also known as arginine dihydrolase) pathway provides carbon, nitrogen, and energy (Weathers et al. 1978). In Synechocystis sp. strain PCC 6803, based on in vivo studies with 14C-labeled substrates and mutational analysis, a model for arginine catabolism was proposed involving an arginase-like pathway combined with a urea cycle that would at the same time degrade aspartate (Quintero et al. 2000). The activity of arginine decarboxylase was also detected, but it was apparently less important than the arginase-like route under the experimental conditions investigated (Quintero et al. 2000). On the other hand, in a large-scale proteomic study of Synechocystis sp. strain PCC 6803, the enzymes of the arginine decarboxylation pathway (arginine decarboxylase and agmatinase) were observed to be up-regulated under certain environmental perturbations (Wegener et al. 2010). The arginine decarboxylase pathway, including conversion of putrescine to succinate, might serve as a source of carbon and nitrogen, as it has been described in Escherichia coli and Pseudomonas sp. (Kurihara et al. 2005; Chou et al. 2008). However, bioinformatic analysis of the genomes of 24 cyanobacteria (including Synechocystis sp. strain PCC 6803) failed to identify genes encoding putrescine oxidase or putrescine transaminase (needed to produce 4-aminobutyraldehyde in the arginine decarboxylase pathway that ends in succinate), suggesting that the arginine decarboxylase pathway in cyanobacteria could be mainly involved in the synthesis of polyamines and the production of ammonium from arginine (Schriek et al. 2007). Additionally, an amino acid oxidase with specificity for basic amino acids (especially arginine) that can release ammonium for growth is present in some cyanobacteria (Flores et al. 1982; Gau et al. 2007). On the other hand, arginine:glycine amidinotransferases involved in secondary metabolism have been described in some cyanobacteria (Muenchhoff et al. 2010; Barón-Sola et al. 2013).
Figure 1.
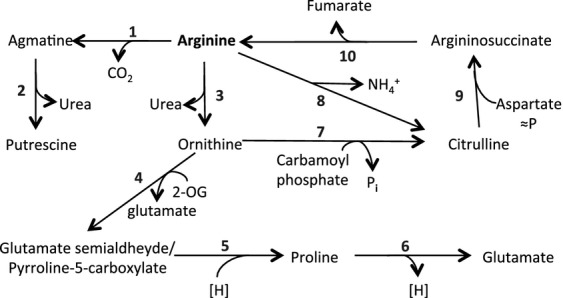
Schematic representation of the arginine decarboxylase, arginase and arginine deiminase pathways. Enzymes and the possible corresponding ORFs in the genome of Anabaena sp. strain PCC 7120 (Kaneko et al. 2001) are as follows: 1, arginine decarboxylase (all3401); 2, agmatinase (alr2310); 3, arginase; 4, ornithine transaminase (alr1080); 5, Δ1pyrroline-5-carboxylate reductase (alr0488); 6, proline oxidase (alr0540); 7, ornithine carbamoyltransferase (alr4907); 8, arginine deiminase; 9, argininosuccinate synthetase (alr4798); 10, argininosuccinate lyase (alr3887). Note that no gene is annotated as encoding arginase or arginine deiminase. 2-OG, 2-oxoglutarate; [H], reducing power; ≈P, energy-requiring reaction (energy provided by the hydrolysis of ATP).
In the Anabaena genome, genes encoding arginine decarboxylase and enzymes corresponding to the second and subsequent steps of the arginase pathway can be identified (Kaneko et al. 2001; see Fig.1). On the other hand, although the presence of an arginase-encoding gene is not evident, open reading frame (ORF) alr2310 is annotated as “similar to agmatinase” (Kaneko et al. 2001). Arginases and agmatinases belong to the ureohydrolase protein family, which has led to wrong genomic annotations in the predicted functions of some ureohydrolase family proteins from different organisms, as has been the case for Synechocystis sp. strain PCC 6803 (Sekowska et al. 2000; see also Quintero et al. 2000). Therefore, it may be asked whether the product of ORF alr2310 is an agmatinase or an arginase. In this work, we created and characterized alr2310 mutants of Anabaena including deletion and reporter-expressing strains. We found that alr2310 encodes an agmatinase that is mainly present in vegetative cells and is required for diazotrophic growth.
Materials and Methods
Strains and growth conditions
Anabaena sp. (also known as Nostoc sp.) strain PCC 7120 was grown axenically in BG11 medium (containing NaNO3), BG110 medium (free of combined nitrogen) or BG110 NH4+ medium (BG110 containing 4 mmol/L NH4Cl and 8 mmol/L 2-[Tris(hydroxymethyl)-methylamino]-ethanesulfonic acid (TES)-NaOH buffer, pH 7.5). In every case, ferric citrate replaced the ferric ammonium citrate used in the original receipt (Rippka et al. 1979). For plates, medium was solidified with 1% separately autoclaved Difco agar. Cultures were grown at 30°C in the light (25 μE m−2 sec−1), with shaking (80–90 rpm) for liquid cultures. Alternatively, cultures (referred to as bubbled cultures) were supplemented with 10 mmol/L of NaHCO3 and bubbled with a mixture of CO2 and air (1% v/v) in the light (50–75 μE m−2 sec−1). For mutants described below, antibiotics were used at the following concentrations: erythromycin (Em), 5 μg mL−1 for liquid cultures and 10 μg mL−1 for solid media; streptomycin sulfate (Sm) and spectinomycin dihydrochloride pentahydrate (Sp), 5 μg mL−1 each for both liquid and solid media; and neomycin sulfate (Nm), 40 μg mL−1 for solid media. DNA was isolated from Anabaena sp. by the method of Cai and Wolk (1990). Anabaena sp. strain FQ163, the hepP mutant (López-Igual et al. 2012), was grown in BG11 medium, supplemented when appropriate with antibiotics. All Anabaena strains used in this work are listed in Table S1.
Escherichia coli DH5α was used for plasmid constructions. It and strains HB101 and ED8654, used for conjugations with Anabaena sp., were grown in Luria–Bertani medium supplemented when appropriate with antibiotics at standard concentrations.
Plasmid construction and genetic procedures
Open reading frame alr2310 of the Anabaena chromosome (Kaneko et al. 2001) was inactivated by deleting an internal fragment of 927 bp. DNA fragments upstream (667 bp) and downstream (501 bp) from the central region of the gene were amplified by standard PCR using as template DNA from Anabaena and primers alr2310-3/alr2310-6 and alr2310-4/alr2310-5 (all oligodeoxynucleotide primers are listed in Table S2). The external primers alr2310-3 and alr2310-4 included SacI-sites in their 5′ ends and primers alr2310-5 and alr2310-6 included EcoRV-sites in their 5′ ends. The upstream and downstream DNA fragments from this gene were cloned in pMBL-T (Dominion MBL, Córdoba, Spain), sequenced, and transferred as a SacI-ended fragment to SacI-digested pCSBN1 (which is a plasmid derived from pCSV3 and pRL278 containing a NmR gene cassette and the sacB gene for positive selection), producing pCSBN5 (all plasmids are listed in Table S1).
Conjugation of Anabaena with E. coli HB101 carrying the cargo plasmid (pCSBN5) with helper and methylation plasmid pRL623 was effected by the conjugative plasmid pRL443, carried in E. coli ED8654, and performed as described (Elhai et al. 1997) with selection for resistance to Nm. Filaments of eight NmR clones were spread on BG110 NH4+ medium supplemented with 5% sucrose (Cai and Wolk 1990), and individual SucR colonies were checked by PCR looking for clones that had substituted the wild-type locus by a deleted locus. The genetic structure of selected clones was studied by PCR with DNA from those clones and primers alr2310-3/alr2310-7 and alr2310-3/alr2310-8. A clone homozygous for the mutant chromosomes was named strain CSMI11 (Δalr2310).
The plasmid carrying fusion gene alr2310-gfp was prepared as follows. A 681-bp fragment from the 3′-terminal part of alr2310 was amplified by PCR using primers alr2310-11 and alr2310-13 (which lacks the stop codon of the gene and contains a sequence encoding a 4-Gly linker and a NheI site in its 5′ end) and Anabaena DNA as template, and the resulting fragment was cloned in vector pMBL-T producing plasmid pCSMI42. This fragment was corroborated by sequencing and transferred as a SacI/NheI-ended fragment to SacI/NheI-digested pCSAL33 (which contains, cloned in vector pMBL-T, the gfp-mut2 gene with an Ala-encoding codon instead of the Met start codon) producing plasmid pCSMI44 that carries the fusion of the gfp gene to the 3′ end of alr2310. The resulting fusion was finally transferred as a KpnI-ended fragment to KpnI-digested pCSV3, which provides resistance to Sm and Sp (Valladares et al. 2011), producing pCSMI46. This plasmid, which bears the alr2310-gfp fusion gene, was transferred to Anabaena by triparental mating as described above, with selection for SmR/SpR. Insertion into alr2310 and segregation of chromosomes carrying the fusion was confirmed by PCR analysis using template DNA from exconjugant clones and primers alr2310-13 and pRL500-1 for testing insertion of gfp-mut2 and alr2310-13 and alr2310-16 for testing segregation of the mutated chromosomes. A homozygous clone bearing the alr2310-gfp construct was named strain CSMI21.
Complementation of the alr2310 deletion mutant was performed with a replicating plasmid. Using DNA from Anabaena as template and primers alr2310-15/alr2310-16, which include SmaI sites close to their 5′ ends, ORF alr2310 was amplified by PCR and ligated into pSpark (Canvax Biotech S.L., Córdoba, Spain) producing plasmid pCSMI53, whose insert was corroborated by sequencing. This insert was excised from pCSMI53 by digestion with SmaI and transferred to SmaI-digested pRL3845, replacing all1711 by alr2310, producing pCSMI54. (pRL3845 is a CmR EmR-plasmid that contains a PglnA-all1711 construct and can replicate in Anabaena; López-Igual et al. 2012.) As is the case for all1711 in pRL3845, alr2310 in pCSMI54 is expressed from the Anabaena glnA promoter. This plasmid was conjugated into strain CSMI11 (Δalr2310) as described above, with selection for EmR, and tested for complementation of the mutant phenotype.
Agmatinase assay in cell-free extracts
Filaments from 800 mL of cultures of Anabaena wild type and strain CSMI11 grown for 5 days in bubbled BG11 medium were harvested and washed with 10 mmol/L N-tris(hydroxymethyl)-methylglycine (Tricine)-NaOH buffer (pH 8.5) and resuspended with 100 mmol/L Tricine-NaOH buffer (pH 8.5) supplemented with 1 mmol/L dithiothreitol, 1 mmol/L MnCl2 and a protease inhibitor mixture tablet (cOmplete Tablets, Mini Ethylenediaminetetraacetic acid (EDTA)-free, Roche, Basel, Switzerland). Filament suspensions were passed twice through a French pressure cell at 20,000 psi. Cell debris was removed by centrifugation at 27,216g, 10 min at 4°C. The resulting supernatant constituted the cell-free extract from vegetative cells.
For determining agmatinase activity, cell-free extracts were supplemented with 5 mmol/L acetohydroxamic acid (a urease inhibitor) and incubated for 30 min at 30°C before adding 1 mmol/L agmatine sulfate (final concentration). The reaction was carried out at 30°C for 180 min. Reactions run without added agmatine were used as controls. Samples of 0.1 mL of the reaction mixture were taken at different times, supplemented with perchloric acid (final concentration, 5%), incubated at 0°C for 10 min and centrifuged at 16,100g, 5 min, at 4°C. The urea produced in the reaction was determined colorimetrically in 0.1 mL of the resulting supernatant by the method of Boyde and Rahmatullah (1980).
Growth tests and nitrogenase activity
Protein concentration and chlorophyll a (Chl) content of the cultures or cell-free extracts were determined by a modified Lowry procedure (Markwell et al. 1978) and by the method of Mackinney (1941), respectively. The growth rate constant (μ = ln2/td, where td is the doubling time) was calculated from the increase of protein content, determined in 0.2 mL samples, of shaken liquid cultures (Montesinos et al. 1995). Cultures were inoculated with an amount of cells containing about 5 μg of protein mL−1 and grew logarithmically until reaching about 40 μg of protein mL−1.
For growth tests in solid media, cultures grown in BG110 NH4+ medium (supplemented with antibiotics when appropriate) were harvested and washed three times with 50 mL of BG110 medium, and dilutions were prepared in BG110 medium. Ten microliter samples of the resulting suspensions were spotted on agar plates with different nitrogen sources and incubated at 30°C in the light (25 μE m−2 sec−1). When indicated, solid media were supplemented with filter-sterilized l-putrescine, agmatine sulfate or l-arginine purchased from Sigma-Aldrich (St. Louis, MO, USA).
Nitrogenase activity was determined by the acetylene reduction assay as described previously (Montesinos et al. 1995). Cells grown in 50 mL of BG110 NH4+ medium were incubated 24 h without combined nitrogen (BG110 medium) under growth conditions and used in the acetylene reduction assays performed under oxic or anoxic conditions. For the latter, the cell suspensions were placed in sealed flasks and supplemented with 10 μmol/L 3-(3,4-dichlorophenyl)-1,1-imethylurea (DCMU), bubbled with argon for 3 min, and incubated for 90 min under assay conditions before starting the reaction by addition of acetylene.
Cyanophycin measurements
To determine cyanophycin, filaments grown in BG110 NH4+ medium were washed with BG110 medium, inoculated at 0.5 μg Chl mL−1 in 50-mL cultures with BG110 NH4+ or BG11 media, and incubated for 8 days under growth conditions. Cyanophycin granule polypeptide (CGP) was isolated from filaments collected from these cultures. The filaments were harvested by centrifugation at 1,935g at room temperature, washed twice with, and resuspended in, sterile double destilled-purified H2O, and disrupted with a French pressure cell (two passages at 20,000 psi). After measuring the obtained volume of cell extract, Chl was determined in a 100-μL sample. The remnant of each extract was centrifuged for 15 min, at 4°C and 27,216g and the pellets were washed twice with 11 mL of sterile milliQ-purified H2O and resuspended in 1 mL of 0.1 mol/L HCl for solubilization of CGP. After 2–4 h of incubation at room temperature and centrifugation under the same conditions, the resulting supernatants were removed and stored at 4°C. The pellets were resuspended in 1 mL of 0.1 mol/L HCl and incubated overnight at room temperature to complete solubilization of CGP, and the solution was subjected to centrifugation as above. The supernatants were combined with those obtained after the first centrifugation and stored at 4°C. These preparations were used for arginine guanidine group determination carried out by the Sakaguchi reaction as modified by Messineo (1966).
Arginine catabolism
Cells grown in BG110 NH4+ medium were harvested by centrifugation at 1,935g at room temperature, washed twice with 25 mmol/L Tricine–NaOH buffer (pH 8.1), and resuspended in the same buffer. The uptake assays were carried out at 30°C in the light (white light from fluorescent lamps, about 175 μE m−2 sec−1) and were started by mixing a suspension of cells (2.1 mL) containing 5–10 μg of Chl mL−1 with a solution (0.1 mL) of l-[U-14C]arginine (274 mCi mmol−1, from PerkinElmer, Waltham, MA, USA). The final concentration of arginine in the experiment was 1 μmol/L. The rate of arginine uptake in the 10- and 30-min assays was estimated by taking a 0.5-mL sample of the cell suspension. The sample was filtered (0.45 μm-pore-size Millipore HA filters were used) and the cells on the filters were washed with 5–10 mL of 5 mmol/L Tricine–NaOH buffer (pH 8.1). The filters carrying the cells were then immersed in 5 mL of scintillation cocktail, and their radioactivity was measured. Retention of radioactivity by boiled cells was used as a blank.
To determine metabolites produced from the labeled arginine at the end of the 10- and 30-min incubations, samples of 0.5 mL of the cell suspension were immediately (<15 sec) mixed, without filtering the cells, with 1.5 mL of water at 100°C and further incubated for 5 min in a bath of boiling water. The resulting suspensions were centrifuged, and samples (1–1.5 mL) from the supernatants were lyophilized and dissolved in 50 μL of water. Samples of the resulting solutions were applied to 0.1-mm-thick cellulose thin-layer chromatography (TLC) plates (20 × 20 cm; Merck, Darmstadt, Germany). Two-dimensional separation of amino acids was effected by using the following solvents: the first dimension solvent consisted of n-butanol–acetone–ammonium hydroxide–water (20:20:10:4, vol/vol/vol/vol), and the second dimension solvent consisted of isopropanol-formic acid-water (20:1:5, vol/vol/vol). The TLC plates were analyzed by electronic autoradiography using a two-dimensional scanner for β particles (Cyclone Plus Phosphor Imager, PerkinElmer, Waltham, MA, USA), which allows a quantitative analysis of the radioactive spots. Identification of the metabolite originating a radioactive spot was made by co-chromatography by supplementing the samples with stable amino acids as markers and visualizing the amino acids after chromatography with a solution of ninhydrin in acetone in the presence of cadmium acetate (Atfield and Morris 1961).
Determination of in vivo protein synthesis
To determine the radioactivity incorporated into macromolecules, a sample of the cell suspension in growth medium was incubated with 10 μmol/L l-leucine supplemented with l-[U-14C]leucine (316 mCi mmol−1, from American Radiolabeled Chemicals, Inc., St. Louis, MO, USA). Samples of 0.5 mL were collected at different times and filtered to determine uptake into whole cells as above. Additionally, samples of 0.5 mL were added to ice-cold trichloroacetic acid (TCA; final concentration, 10%), incubated in ice-water for 60 min, and filtered as above. The filters were washed with 5 mL of ice-cold 10% TCA and immersed in a scintillation cocktail, and their radioactivity was measured.
Western blot analysis
Filaments from 100 mL of cultures of Anabaena sp. strains PCC 7120 and CSMI21 grown in bubbled BG11 and BG110 media were harvested, washed with buffer 1 (50 mmol/L Tricine-HCl buffer, pH 7.5, 10% glycerine, 100 mmol/L NaCl), and resuspended in 4 mL of the same buffer. To isolate heterocysts, 800 mL of cultures grown for 48 h in bubbled BG110 medium were harvested and washed with buffer 2 (50 mmol/L imidazol and 0.5 mmol/L EDTA, pH 8.0), resuspended in 10 mL of the same buffer, incubated with 1 mg mL−1 of lysozyme for 15 min at room temperature and centrifuged at 1935g (4°C, 10 min). The pellets were resuspended in 5 mL of buffer 2 and passed three times through a French pressure cell at 3,000 psi. Samples were enriched in heterocysts after successive steps of low-speed centrifugation (200g, 10 min, 4°C) and washing with buffer 1, and the isolated heterocysts were resuspended in 4 mL of buffer 1. The filament and heterocyst suspensions were both supplemented with a protease inhibitor cocktail tablet (cOmplete Tablets, Mini EDTA-free, Roche) and passed two times through a French pressure cell at 20,000 psi. Cell debris was removed by centrifugation at 16,100g (4°C, 10 min). Proteins in the supernatants were resolved by SDS-PAGE and transferred to Hybond-P PVDF membranes (GE Healthcare, Buckinghamshire, England). Western analysis was performed incubating with an anti-GFP antibody (A6455 from Invitrogen, Carlsbad, CA, USA) diluted 1:2000 in Tween 20, Tris-buffered saline (TTBS) with 5% non-fat milk powder. Antigen–antibody complexes were visualized with a peroxidase-conjugated anti-rabbit-IgG antiserum (Sigma-Aldrich, St. Louis, MO, USA) and developed with the WesternBright™ Western blotting detection kit (Advansta, Menlo Park, CA, USA). A cell-free extract from Anabaena carrying pAM1954 (Yoon and Golden 1998) was used as a free GFP-containing control.
Microscopy
Cells grown during 5–7 days in shaken liquid BG110 NH4+ medium or heterocyst preparations were observed and photographed with a Zeiss (Oberkochen, Germany) Axioscop microscope equipped with a Zeiss ICc1 digital camera. GFP fluorescence was analyzed by confocal microscopy. Samples from cultures of Anabaena sp. strain CSMI21 or the wild-type PCC 7120 as a control grown in bubbled cultures with BG11 or BG110 medium were visualized using a Leica HCX PLAN-APO 63X 1.4 NA oil immersion objective attached to a Leica TCS SP2 confocal laser-scanning microscope. GFP was excited using 488-nm irradiation from an argon ion laser. Fluorescence emission was monitored by collection across windows of 500–520 nm (GFP imaging) and 630–700 nm (cyanobacterial autofluorescence). Under the conditions used, optical section thickness was about 0.4 μm. GFP fluorescence intensity was analyzed using ImageJ 1.43 m software (http://imagej.nih.gov/ij/).
Results
Construction and phenotype of an alr2310 mutant
The Anabaena ORF alr2310 is located between alr2309 encoding a single-stranded nucleic acid binding protein and alr2311 encoding an RNA-binding protein (Fig.2). Therefore, there is no obvious functional relation between the product of alr2310 and those of its surrounding genes, and there is no evidence for co-transcription of these genes (Flaherty et al. 2011). Nonetheless, to investigate the role of the alr2310 avoiding any possible polar effects of the mutation, a 927-bp in-frame internal fragment of the gene was removed without leaving any genetic marker in it (Fig.2A; see Fig. S1 and Materials and Methods for details). The Δalr2310 mutant, strain CSMI11, was homozygous for mutant chromosomes (Fig. S1). Tests in solid media showed that strain CSMI11 is impaired in growth in the presence of combined nitrogen (ammonium or nitrate) and is unable to grow under diazotrophic conditions (Fig.2B). In shaken liquid cultures, the growth rate constant of strain CSMI11 in ammonium- or nitrate-containing medium was, respectively, similar to or somewhat lower than that of the wild type, whereas the growth rate constant in combined N-free media was about 9% that of the wild type (Table1), confirming that strain CSMI11 is hampered in diazotrophic growth. Complementation of CSMI11 with a plasmid bearing wild-type alr2310 allowed diazotrophic growth, corroborating that growth impairment resulted from the alr2310 mutation (see strain CSMI11-C in Fig.2B). After incubation without a source of combined nitrogen, strain CSMI11 produced heterocysts that appeared immature (Fig. S2) and showed no nitrogenase activity, measured under both oxic and anoxic conditions (Table1). The production of aberrant heterocysts that lack nitrogenase activity can explain the impairment of strain CSMI11 in diazotrophic growth.
Figure 2.
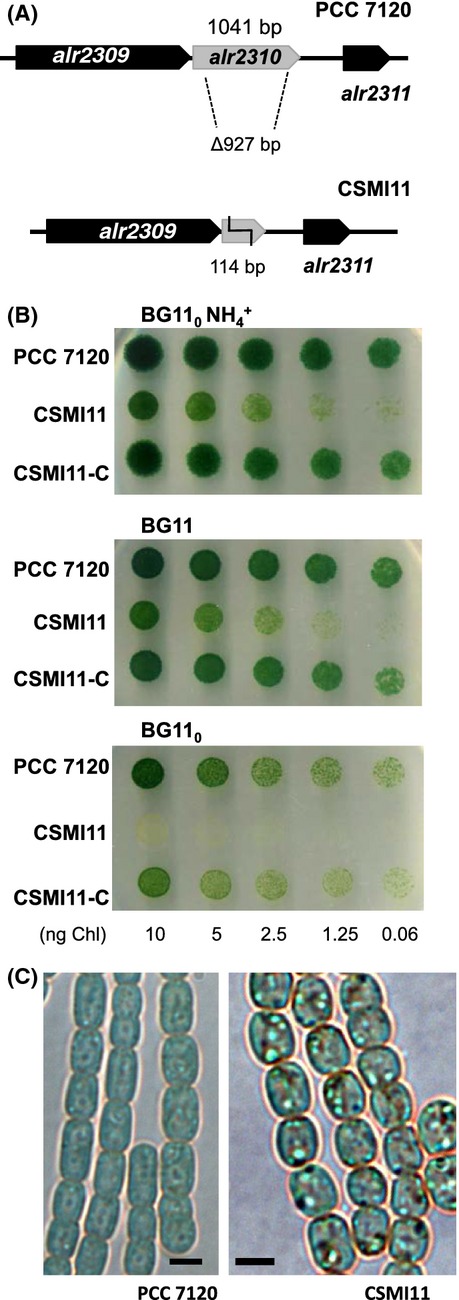
Characterization of an Anabaena alr2310 mutant. (A) Schematic of the alr2310 genomic region in Anabaena with indication of the DNA fragment removed to create strain CSMI11. (B) Growth tests in solid medium using ammonium (BG110 NH4+), nitrate (BG11) or N2 (BG110) as the nitrogen source. Each spot was inoculated with an amount of cells containing the indicated amount of Chl, and the plates were incubated under culture conditions for 7 days and photographed. Strain CSMI11-C is strain CSMI11 complemented with alr2310. (C) Filaments of Anabaena wild type (PCC 7120) and strain CSMI11 from cultures incubated for 5 days in BG11 medium and visualized by light microscopy. Scale bars, 2 μm.
Table 1.
Growth rate constants, nitrogenase activity, and cyanophycin granule polypeptide (CGP) levels in Anabaena sp. strains PCC 7120 (wild type) and CSMI11 (Δalr2310).
| Strain | Growth rate, μ (day−1) | Nitrogenase activity (μmol [mg Chl]−1 h−1) | CGP (μg arginine [mg Chl]−1) | ||||
|---|---|---|---|---|---|---|---|
| NH4+ | NO3− | N2 | Oxic | Anoxic | NH4+ | NO3− | |
| PCC 7120 | 0.55 ± 0.08 (4) | 0.79 ± 0.12 (8) | 0.67 ± 0.22 (9) | 7.13 ± 2.58 (3) | 15.26 ± 3.05 (5) | 115.12 ± 85.80 (3) | 147.06 ± 6.61 (3) |
| CSMI11 | 0.56 ± 0.11 (4) | 0.62 ± 0.12 (4) | 0.06 ± 0.04 (4) | 0.00 ± 0.00 (4) | 0.003 ± 0.003 (3) | 516.71 ± 123.89 (3) | 1211.11 ± 429.58 (3) |
| t test | P = 0.90 | P = 0.06 | P = 0.0003* | P = 0.005* | P = 0.0003* | P = 0.02* | P = 0.02* |
The growth rate constant (μ) was determined in photoautotrophic shaken cultures with the indicated nitrogen source. To determine nitrogenase activity, filaments grown in BG110 NH4+ medium and incubated in nitrogen-free BG110 medium for 24 h were used in assays of reduction of acetylene to ethylene under oxic and anoxic conditions. Cyanophycin was determined by the Sakaguchi reaction for arginine on CGP granules isolated from ammonium- or nitrate-grown filaments. Figures are the mean and SD of data from the number of independent experiments indicated in parenthesis. The significance of the differences between the mutant and the wild-type figures was assessed by the Student's t test (P indicated in each case); asterisks denote likely significant differences.
Microscopic examination of strain CSMI11 showed the presence of abundant granulation in the cytoplasm of the cells growing in the presence of nitrate or ammonium (shown in Fig.2C for nitrate-grown filaments). To test whether that granulation corresponded to cyanophycin, CGP isolation was carried out and the isolated material was measured with the Sakaguchi reaction for arginine. CSMI11 cells grown for 8 days in the presence of nitrate or ammonium had, respectively, about 8.3- and 4.5-fold higher amounts of CGP than the control wild type cells (Table1).
Arginine catabolism
Wild-type Anabaena and strain CSMI11 were used in uptake assays with [14C]arginine, and the fate of arginine was studied by TLC analysis of the radiolabeled compounds produced in filaments that had been incubated in BG11 or BG110 medium. Consistent with results previously published for Anabaena (Montesinos et al. 1995), arginine was taken up at appreciable levels in filaments from either BG11 or BG110 medium, and uptake was significant in both mutant CSMI11 and the wild type (Table2). Arginine was metabolized in both strains. After 30 min of incubation the amount of radioactivity that remained associated to arginine accounted for only 21.6 and 19.2% in BG11-grown cells of the wild type and the mutant, respectively, and for 50.3 and 17.4% in cells of the corresponding strain that had been incubated in BG110 medium (Table2). Because metabolism was observed to proceed to a somewhat larger extent in mutant CSMI11 than in the wild type, catabolism of arginine is not inhibited by the inactivation of alr2310.
Table 2.
l-[14C]arginine uptake and metabolic products in Anabaena sp. strains PCC 7120 (wild type) and CSMI11 (Δalr2310).
| BG11 | BG110 | |||||||
|---|---|---|---|---|---|---|---|---|
| PCC 7120 | CSMI11 | PCC 7120 | CSMI11 | |||||
| 10 min | 30 min | 10 min | 30 min | 10 min | 30 min | 10 min | 30 min | |
| Arginine taken up (nmol·[mg Chl]−1) | 73.3 | 115.9 | 57.3 | 72.4 | 93.7 | 162.3 | 91.3 | 124.5 |
| Labeled compounds (%) | ||||||||
| Origin | 7.5 | 18.6 | 1.8 | 8.4 | 10.0 | 12.4 | 10.2 | 14.2 |
| Arginine | 54.8 | 21.6 | 52.6 | 19.2 | 66.7 | 50.3 | 51.1 | 17.4 |
| Agmatine | 0.4 | 0.4 | 2.9 | 10.4 | 0.2 | 0.3 | 7.1 | 15.4 |
| Aspartate | 1.5 | 1.5 | 2.1 | 1.8 | 0.5 | 0.8 | 1.1 | 1.2 |
| Glutamate | 7.0 | 33.0 | 24.6 | 52.0 | 1.6 | 10.7 | 8.6 | 37.1 |
| Glutamine/Citrulline | 6.4 | 3.2 | 3.7 | 1.2 | 2.5 | 2.0 | 1.8 | 1.1 |
| Ornithine | 2.2 | 1.0 | 1.6 | 1.2 | 2.5 | 1.6 | 2.4 | 1.5 |
| Proline | 15.4 | 17.4 | 8.6 | 2.7 | 9.5 | 15.8 | 9.3 | 6.4 |
| Spot #6 | 4.6 | 2.5 | 2.1 | 1.5 | 6.6 | 5.1 | 8.4 | 3.3 |
| Spot #7 | – | 0.8 | – | 1.7 | – | 1.3 | – | 2.4 |
Filaments of the indicated strains grown in BG110 NH4+ medium and incubated for 24 h in BG11 or BG110 medium were used at 5–10 μg Chl mL−1 in assays of uptake of 1 μmol/L l-[14C]arginine as described in Materials and Methods. After 10 and 30 min of incubation, uptake of arginine was determined and metabolites in the suspensions were analyzed by TLC. Cell-associated metabolites are presented as the percentage of the sum of radioactivity in 14C-labeled chromatographic spots after subtraction of extracellular arginine. Compounds that do not move with the used solvents accumulate in the origin of the chromatography.
In the suspensions of wild-type filaments from BG11 medium (Fig.3, Table2), after 10 min of incubation, radioactivity from [14C]arginine was mainly distributed among proline, glutamate, glutamine/citrulline (which overlap in the TLC system of solvents used in this study), ornithine, aspartate, and unidentified spot #6. After 30 min, an additional unidentified spot, #7, became visible, and a notable accumulation of radioactivity in glutamate and proline was observed. In strain CSMI11, the general pattern of labeled compounds was similar to that observed in the wild type but, remarkably, a spot that could be identified as agmatine was detected (Fig.3) that accounted for a significant 10.4% of the cell-associated radioactivity after 30 min of incubation. Additionally, substantially more radioactivity accumulated in glutamate, and less in proline, in strain CSMI11 than in the wild type.
Figure 3.
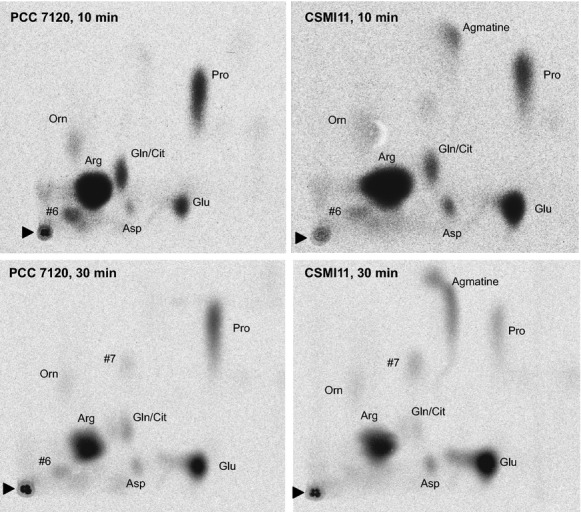
Production of 14C-labeled metabolites from l-[14C]arginine in filaments of Anabaena wild type (PCC 7120) and strain CSMI11 grown in BG110 NH4+ medium and incubated in BG11 (nitrate-containing) medium for 24 h. Suspensions of filaments containing 5–10 μg of Chl mL−1 were incubated for 10 and 30 min with 1 μmol/L l-[14C-(U)]arginine. Metabolites in the cell suspensions were extracted and analyzed by TLC and autoradiography as described in Materials and Methods. The amino acids identified were as follows: arginine (Arg), citrulline (Cit), proline (Pro), glutamate (Glu), glutamine (Gln), ornithine (Orn), aspartate (Asp), and agmatine. Two unidentified spots, indicated as #6 and #7, were also detected. Note that the glutamine and citrulline spots overlap. The black triangles point to the origin of the chromatography.
In the suspensions of filaments that had been incubated in BG110 medium, the patterns of [14C]arginine-derived products were similar to those found in the filaments from BG11 medium, although the accumulation of radioactivity in aspartate and, especially, in glutamate was lower in the filaments of both strains from BG110 medium (Fig. S3; Table2). However, in the filaments from BG110 medium, the accumulation of radioactivity in glutamate was also higher in the mutant than in the wild type (Table2). Additionally, a specific accumulation of agmatine, accounting for 15.4% of the cell-associated radioactivity, was also detected in filaments of the mutant from BG110 medium (Fig. S3).
The results showing that agmatine accumulates specifically in mutant CSMI11 suggest that alr2310 is an agmatinase. We therefore determined agmatinase in cell-free extracts from BG11-grown filaments as production of urea from agmatine. Whereas an activity of about 10 nmol urea produced (mg Chl)−1 h−1 was detected in extracts from the wild type, the activity was undetectable in extracts from strain CSMI11.
Growth tests with arginine and arginine catabolism-related compounds
Alterations in growth of the Δalr2310 mutant, strain CSMI11 (lacking the activity of reaction 2 in Fig.1), could be explained by a lack of putrescine, which might be required as a polyamine or for production of other polyamines, or by the accumulation of agmatine, which might be detrimental to the cells. The effects of putrescine and agmatine on Anabaena were assessed by growth tests in solid medium, which were performed at a fixed pH of 7.5. Growth tests with different nitrogen sources and l-putrescine at 100 or 300 μmol/L were performed, but no (or a very poor) recovery of the growth of strain CSMI11 in BG110 medium was observed (Fig. S4). Although we cannot rule out that putrescine is not taken up by Anabaena, this possibility is unlikely because putrescine uptake has been reported in several cyanobacteria (Guarino and Cohen 1979; Raksajit et al. 2006) and the Anabaena genome bears genes encoding a putative polyamine ABC-type transporter (Kaneko et al. 2001).
To test whether the impairment of strain CSMI11 in diazotrophic growth could result from an accumulation of agmatine, growth tests with different nitrogen sources and agmatine were performed. Whereas good growth was observed in the presence of 10 or 100 μmol/L agmatine (not shown), 1 mmol/L agmatine was inhibitory for both the wild type and strain CSMI11, although in the presence of nitrate or ammonium the mutant was more sensitive than the wild type (Fig.4). The higher sensitivity of the mutant can be related to its inability to metabolize agmatine. In addition, these results show that the degree of agmatine toxicity depends on the nitrogen source.
Figure 4.
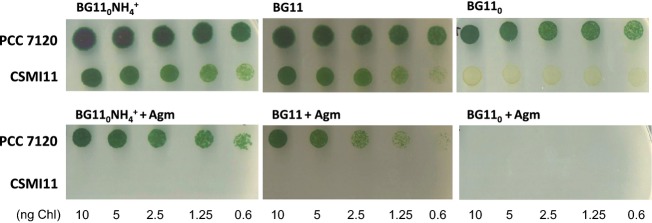
Growth test of Anabaena wild type (PCC 7120) and strain CSMI11 in BG110 NH4+-, BG11-, and BG110-solid media supplemented with 10 mM TES-NaOH buffer (pH 7.5) and, when indicated, 1 mmol/L agmatine sulfate (Agm). Each spot was inoculated with an amount of cells containing the indicated amount of Chl, and the plates were incubated for 8 days under culture conditions and photographed.
To assess the possibility that agmatine is competing with arginine in cellular metabolism, we investigated whether the growth defect of strain CSMI11 in media lacking combined nitrogen could be rescued by arginine. Strain FQ163, a hepP gene mutant that is unable to grow fixing N2 under oxic conditions (Fox− phenotype; López-Igual et al. 2012), was used as a control. Supplementation of the medium with 0.1–30 mmol/L arginine rescued the growth of both strains, CSMI11 and FQ163, but growth was stronger in the case of FQ163 than in the case of CSMI11 (Fig.5). Arginine can be utilized as a nitrogen source by Anabaena (Herrero and Flores 1990; Burnat et al. 2014), and it clearly serves as a nitrogen source for the Fox− strain FQ163. In strain CSMI11, however, arginine compensates only to a limited extent the effect of inactivation of alr2310. It is possible that agmatine produced from arginine contributes to hamper growth.
Figure 5.
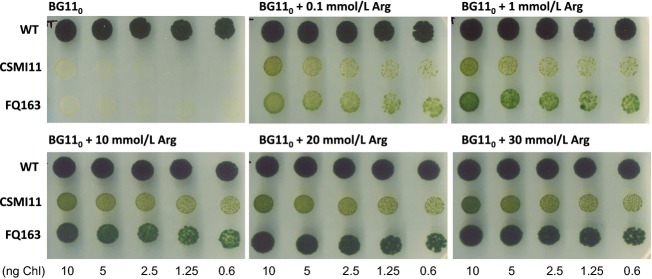
Growth tests of Anabaena sp. strains PCC 7120 (WT), CSMI11 (Δalr2310) and FQ163 (hepP) in BG110 solid medium supplemented with 10 mmol/L TES-NaOH buffer (pH 7.5) and, when indicated, arginine (Arg) at the specified concentration. Spots were inoculated with an amount of cells containing the indicated amount of Chl, and the plates were incubated for 14 days under culture conditions and photographed. Strain FQ163 was used as a Fox− control.
Protein synthesis in strain CSMI11
To determine if strain CSMI11 could be affected in protein synthesis, radioactivity incorporated from [14C] leucine into macromolecules was assessed. Leucine is an amino acid of low pool size that is primarily incorporated into protein, which makes it an appropriate amino acid to study protein synthesis in vivo (Padan et al. 1971). Cultures of wild-type Anabaena and strain CSMI11 were grown in BG110 NH4+ medium, incubated for 24 h in BG110 or BG11 medium and analyzed. As shown in Table3, leucine uptake was about 1.8-fold higher in strain CSMI11 than in the wild type when incubated in BG11 medium, whereas the rates of uptake were similar for both strains in BG110 medium. Incorporation of radioactivity from [14C]leucine into TCA-precipitable material was 1.6-fold and 0.72-fold in strain CSMI11 as compared to the wild type when incubated in BG11 or BG110 medium, respectively. Because the amount of [14C]leucine that is incorporated into protein can be expected to depend on the amount of [14C]leucine taken up, we compared the fraction of [14C]leucine taken up that was incorporated into TCA-precipitable material in the mutant and the wild type in each of the two growth conditions. Relative incorporation into TCA-precipitable material was about 13% lower in the mutant than in the wild type in BG11 medium, and about 25% lower in BG110 medium. Therefore, the hampered diazotrophic growth of strain CSMI11 is hardly only a consequence of a low level of protein synthesis.
Table 3.
[14C]Leucine uptake and incorporation into TCA-precipitable material in Anabaena sp. strains PCC 7120 (wild type) and CSMI11 (Δalr2310).
| Growth conditions | Strain | [14C]Leucine incorporation (nmol (mg Chl)−1 min−1) | TCA-precipitable material/uptake | |
|---|---|---|---|---|
| Uptake | TCA-precipitable material | |||
| BG11 | PCC 7120 | 14.00 ± 8.59 | 11.91 ± 7.59 | 0.851 |
| CSMI11 | 25.03 ± 17.53 | 18.52 ± 13.43 | 0.740 | |
| BG110 | PCC 7120 | 41.50 ± 28.49 | 25.48 ± 14.07 | 0.614 |
| CSMI11 | 39.97 ± 32.40 | 18.44 ± 14.12 | 0.461 | |
To determine leucine uptake (incorporation into whole cells) and the radioactivity incorporated into macromolecules, suspensions of filaments from cultures with BG11 or BG110 medium were incubated in the same media with 10 μmol/L l-[14C]leucine for 1 h, as described in Materials and Methods. At several time points, samples were filtered to determine leucine uptake rates or mixed with ice-cold TCA and then filtered to determine rates of incorporation into macromolecules. Figures are the mean ± SD of four independent experiments. For the fraction of radioactivity taken up that was precipitable with TCA, Student's t tests indicated that the differences between the mutant and the wild type were likely significant in both BG11 (P = 0.006) and BG110 (P = 0.008) medium.
Cell-specific expression of Alr2310
To investigate the expression and cell localization of Alr2310, an alr2310-gfp fusion gene was constructed and transferred to Anabaena (Fig. S5). Anabaena clones bearing this construct as the only alr2310 gene were readily isolated. Strain CSMI21, which was selected for further analysis, exhibited growth properties similar to those of the wild type (Fig. S5), indicating that the Alr2310-GFP fusion protein retained Alr2310 function.
When strain CSMI21 was grown with nitrate as the nitrogen source, GFP fluorescence was observed in all the cells of the filament, but when incubated in the absence of combined nitrogen, GFP fluorescence was observed in vegetative cells at higher levels than in heterocysts (Fig.6A). Quantification of the GFP fluorescence in vegetative cells and heterocysts indicated that in filaments incubated for 24 or 48 h without combined nitrogen, heterocysts had on average 46.7% and 12.6%, respectively, of the fluorescence detected in vegetative cells (Fig.6B).
Figure 6.
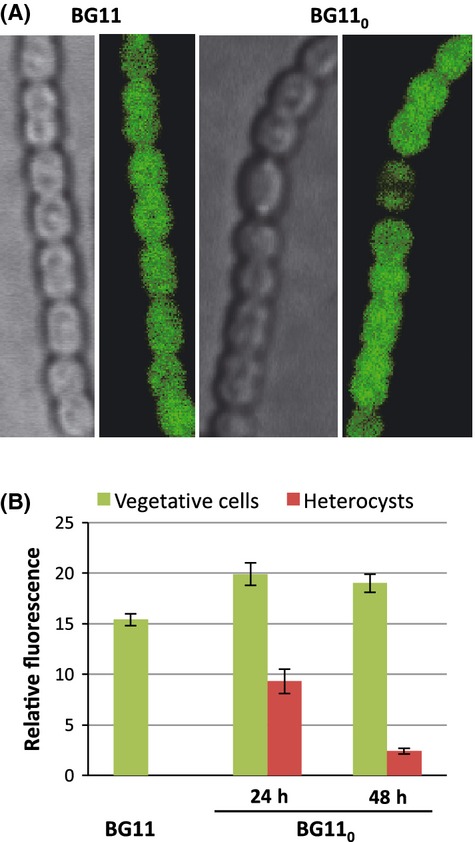
Cellular localization of Alr2310-GFP in the Anabaena filaments. (A) Filaments of Anabaena sp. strain CSMI21 grown in bubbled BG11 medium or incubated in bubbled BG110 medium (without combined nitrogen) for 48 h were visualized by confocal microscopy as described in Materials and Methods. Bright field and GFP fluorescence images are shown Brightness and contrast were increased to improve visibility. (B) Quantification of GFP fluorescence in cells of strain CSMI21. Average background fluorescence from wild-type cells (which lack GFP) was subtracted. Figures are the mean and standard deviation of the mean of the fluorescence recorded in cells grown in bubbled BG11 medium (487 cells counted) or incubated in bubbled BG110 medium for 24 h (309 vegetative cells and 58 heterocysts counted; Student's t test P = 2.7 × 10−18) or 48 h (409 vegetative cells and 67 heterocysts counted; P = 1.3 × 10−56).
Western blot analysis with anti-GFP antibodies identified a band corresponding to the Alr2310-GFP fusion protein in cell-free extracts from filaments of strain CSMI21 grown in bubbled BG11 medium or incubated in bubbled BG110 medium for 48 h (Fig.7). Only a very low amount of free GFP released from the fusion protein was observed. Noteworthy, the fusion protein was not detected in cell-free extracts from heterocysts of strain CSMI21. The results of the western blot analysis are consistent with those of the GFP fluorescence analysis showing that the Alr2310-GFP fusion protein is present at significantly lower levels in heterocysts than in vegetative cells.
Figure 7.
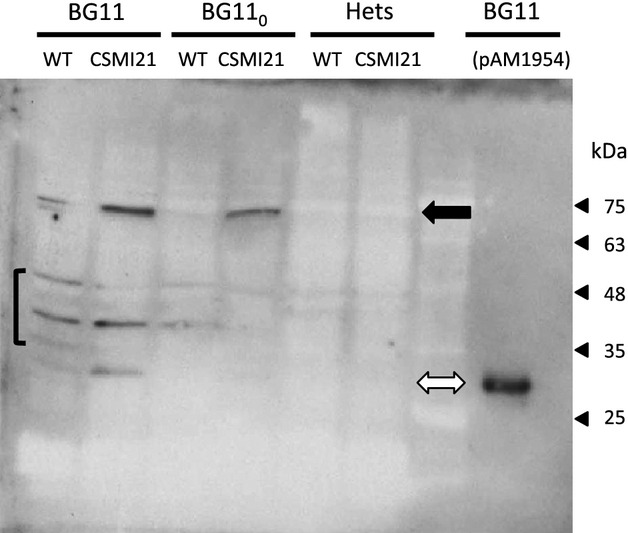
Western blot analysis of the GFP in strain CSMI21 and control strains. The Anabaena strains used were PCC 7120 (WT), mutant CSMI21 (alr2310-gfp) and a strain carrying pAM1954, which is a replicative plasmid bearing the gfp gene expressed from the rbc gene promoter. Cell-free extracts were prepared from whole filaments grown in bubbled cultures with BG11 medium or incubated in BG110 medium for 48 h, or from heterocysts (Hets) isolated from the latter. Anabaena sp. [pAM1954] was grown in shaken cultures containing BG11 medium. Proteins were subjected to SDS-PAGE electrophoresis and transferred to membranes. Protein detection was performed with anti-GFP antibodies. Native GFP is 26.9 kDa and the Alr2310-GFP fusion, which includes Alr2310, a tetra-glycine linker peptide, and the GFP is 65.7 kDa. Protein loaded was 40 μg per well (0.6 μg in the case of Anabaena [pAM1954]). As observed with the wild-type extracts, the antibodies marked 3 or 4 unspecific bands (brackets). Strain CSMI21 produced mainly a band corresponding to the fusion protein (black arrow) and only a faint band corresponding to free GFP, which was identified with the extract of Anabaena [pAM1954] that only produced, as expected, free GFP (open arrow). Note that some material from lane BG11/CSMI21 may have contaminated lane BG11/WT.
Discussion
ORF alr2310 from the genome of the heterocyst-forming cyanobacterium Anabaena putatively encodes an ureohydrolase family protein. Based on bioinformatic analysis, Alr2310 has been proposed to be the arginase of Anabaena (Schriek et al. 2007). However, as demonstrated by the accumulation of agmatine from [14C]arginine in the Δalr2310 mutant, strain CSMI11, and from lack of agmatinase activity in cell-free extracts from this mutant, alr2310 encodes an agmatinase and therefore is the speB gene of Anabaena. Two possible agmatinases, Sll0228 and Sll1077, are encoded in the genome of Synechocystis sp. strain PCC 6803, and Alr2310 is most similar to Sll0228 (Schriek et al. 2007) whose inactivation also leads to lack of any agmatinase activity in Synechocystis cell-free extracts (Quintero et al. 2000). Accumulation of agmatine in strain CSMI11 in turn implies the operation of arginine decarboxylase, the product of ORF all3401, in Anabaena (Fig.1). We have not consistently detected in the wild type a TLC spot that could be identified as putrescine. We do not know, however, whether in our experiments the radioactivity from putrescine could pass to other metabolites or whether our methodology is inadequate to extract this polyamine.
Cells of the CSMI11 mutant grown with combined nitrogen (nitrate or ammonium) show an extensive granulation in the cytoplasm (Fig.1C) that correlates with the accumulation of CGP (Table1). It has been reported that the end products of cyanophycin mobilization, arginine and aspartate, inhibit cyanophycinase (Gupta and Carr 1981a). Agmatine is known to inhibit moderately cyanophycin synthetase (Aboulmagd et al. 2001), but we do not know whether it could also inhibit cyanophycinase as to account for the observed accumulation of CGP in the CSMI11 mutant. Alternatively, if, as is the case for amino acids such as lysine, glutamate, citrulline and ornithine (Merrit et al. 1994; Ziegler et al. 1998; Berg et al. 2000; Aboulmagd et al. 2001), agmatine were incorporated into cyanophycin at significant levels, accumulation of CGP could result from an inefficient degradation of agmatine-containing cyanophycin.
Strain CSMI11 is impaired in diazotrophic growth and produces only immature heterocysts, and CSMI11 filaments incubated for 24 h without combined nitrogen do not show measurable nitrogenase activity (Table1). Inactivation of alr2310 likely results in accumulation of agmatine (as observed with [14C] arginine) and a lack of putrescine, but growth inhibition appears to be related specifically to accumulation of agmatine. Inhibition of the growth of some bacteria by agmatine has previously been noted, and competitive inhibition of amino acid transport or interference with translation were considered as possible mechanisms of inhibition (Griswold et al. 2006). Agmatine may compete with arginine for charging tRNAs resulting in a general inhibition of protein synthesis. We have found that incorporation of [14C]leucine into macromolecules is somewhat inhibited in strain CSMI11 as compared to the wild type, especially in cells that had been incubated in the absence of combined nitrogen, but the observed effect seems insufficient to prevent growth. Because diazotrophic growth of Anabaena requires heterocyst differentiation and, as mentioned earlier, strain CSMI11 produces only immature heterocysts, interference of agmatine with heterocyst differentiation may contribute to prevent growth of this mutant in the absence of combined nitrogen.
Because the pattern of [14C]arginine metabolic products in Anabaena is very similar to that reported for Synechocystis sp. strain PCC 6803 (Quintero et al. 2000), it is possible that the arginine catabolism pathway proposed for this cyanobacterium, which combines the arginase catabolic route and a urea cycle generating proline, glutamate and glutamine as final products (Quintero et al. 2000), is operative also in Anabaena. An interesting aspect of this pathway is that it incorporates aspartate at the level of the urea cycle (Fig.1), thus providing a rationale for the combined utilization of the amino acids released in cyanophycin mobilization, arginine and aspartate. Appreciable levels of arginase activity have been detected in cell-free extracts of Anabaena (Gupta and Carr 1981b), but the gene encoding an enzyme with arginase activity remains to be identified in this cyanobacterium. The possibility that Alr2310 has also arginase activity is not supported by the results of inactivation of alr2310, which does not prevent production of proline and glutamate from [14C]arginine. Instead, production of glutamate from [14C]arginine is significantly increased in mutant CSMI11 as compared to the wild type (Table2). This result suggests that if the arginine decarboxylase pathway is not operative, arginine degradation through the arginase-like pathway is increased. A small amount of [14C]aspartate produced from [14C]arginine could also be observed in our experiments, but we do not know the possible pathway involved in this conversion.
As described in the Introduction, we have recently found that in the diazotrophic filament of Anabaena, the second step of cyanophycin degradation, hydrolysis of β-aspartyl-arginine, takes place mainly in vegetative cells (Burnat et al. 2014). This dipeptide appears to be an important vehicle for the transfer of nitrogen from heterocysts to vegetative cells, consistent with the possible existence of a gradient of arginine or an arginine-containing compound in the diazotrophic filament (Ke and Haselkorn 2013). These results imply an active catabolism of arginine (and aspartate) in the vegetative cells of the diazotrophic filament. Consistently, arginase and a low level of arginine deiminase activity detected in Anabaena are present at significantly higher levels in vegetative cells than in heterocysts (Gupta and Carr 1981b). Our results showing the presence of the Alr2310-GFP fusion protein at significantly higher levels in vegetative cells than in heterocysts indicate that also agmatinase, and hence the arginine decarboxylase pathway, is mainly operative in the vegetative cells of the diazotrophic filament. These observations highlight the importance of arginine catabolism to make available in the vegetative cells the nitrogen transiently stored as cyanophycin and transferred from the heterocysts.
Acknowledgments
We thank Antonia Herrero for useful discussions and Bárbara Nocea and Rocío López-Igual for constructing plasmids pCSBN1, pCSBN4, and pCSBN5. M. B. was the recipient of an FPI fellowship/contract from the Spanish Government. Research was supported by grant no. BFU2011-22762 from Plan Nacional de Investigación, Spain, co-financed by FEDER.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Construction and verification of mutant CSMI11.
Microscopic observation of filaments of mutant strain CSMI11.
Catabolism of L-[14C]arginine in filaments of Anabaena wild type and strain CSMI11 incubated in medium lacking combined nitrogen.
Growth tests of Anabaena wild type and strain CSMI11 in solid media supplemented with putrescine.
Construction, verification and growth test of strain CSMI21.
Cyanobacterial strains and plasmids used in this work.
Oligodeoxynucleotide primers used in this work.
References
- Aboulmagd E, Oppermann-Sanio FB. Steinbüchel A. Purification of Synechocystis sp. strain PCC6308 cyanophycin synthetase and its characterization with respect to substrate and primer specificity. Appl. Environ. Microbiol. 2001;67:2176–2182. doi: 10.1128/AEM.67.5.2176-2182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atfield GN. Morris JOR. Analytical separations by high-voltage paper electrophoresis. Amino acids in protein hydrolysates. Biochem. J. 1961;81:606–614. doi: 10.1042/bj0810606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barón-Sola A, Gutiérrez-Villanueva MA, Del Campo FF. Sanz-Alférez S. Characterization of Aphanizomenon ovalisporum amidinotransferase involved in cylindrospermopsin synthesis. MicrobiologyOpen. 2013;2:447–458. doi: 10.1002/mbo3.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H, Ziegler K, Piotukh K, Baier K, Lockau W. Volkmer-Engert R. Biosynthesis of the cyanobacterial reserve polymer multi-L-arginyl-poly-L-aspartic acid (cyanophycin): mechanism of the cyanophycin synthetase reaction studied with synthetic primers. Eur. J. Biochem. 2000;267:5561–5570. doi: 10.1046/j.1432-1327.2000.01622.x. [DOI] [PubMed] [Google Scholar]
- Boyde TRC. Rahmatullah M. Optimization of conditions for the colorimetric determination of citrulline, using diacetyl monoxime. Anal. Biochem. 1980;107:424–431. doi: 10.1016/0003-2697(80)90404-2. [DOI] [PubMed] [Google Scholar]
- Burnat M, Herrero A. Flores E. Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. USA. 2014;111:3823–3828. doi: 10.1073/pnas.1318564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y. Wolk CP. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr NG. Biochemical aspects of heterocyst differentiation and function. In: Papageorgiou GC, Packer L, editors. Photosynthetic prokaryotes: cell differentiation and function. New York, NY: Elsevier; 1983. pp. 265–280. [Google Scholar]
- Chou HT, Kwon DH, Hegazy M. Lu CD. Transcriptome analysis of agmatine and putrescine catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2008;190:1966–1975. doi: 10.1128/JB.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunin R, Glansdorff N, Piérard A. Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatti L, Flores E. Salerno E. Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 2002;513:175–178. doi: 10.1016/s0014-5793(02)02283-4. [DOI] [PubMed] [Google Scholar]
- Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E. Wolk CP. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997;179:1998–2005. doi: 10.1128/jb.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty BL, van Nieuwerburgh F, Head SR. Golden JW. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics. 2011;12:332. doi: 10.1186/1471-2164-12-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E. Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 2010;8:39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- Flores E, Herrero A. Guerrero MG. Production of ammonium dependent on basic L-amino acids by Anacystis nidulans. Arch. Microbiol. 1982;131:91–94. [Google Scholar]
- Gau AE, Heindl A, Nodop A, Kahmann U. Pistorius EK. L-amino acid oxidases with specificity for basic L-amino acids in cyanobacteria. Z. Naturforsch. 2007;62c:273–284. doi: 10.1515/znc-2007-3-419. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Turner S, Olsen GJ, Barns S, Lane DJ. Pace RN. Evolutionary relationships among cyanobacteria and green chloroplasts. J. Bacteriol. 1988;170:3584–3592. doi: 10.1128/jb.170.8.3584-3592.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AR, Jameson-Lee M. Burne RA. Regulation and physiologic significance of agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 2006;188:834–841. doi: 10.1128/JB.188.3.834-841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino LA. Cohen SS. Mechanism of toxicity of putrescine in Anacystis nidulans. Proc. Natl. Acad. Sci. USA. 1979;76:3660–3664. doi: 10.1073/pnas.76.8.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. Carr NG. Enzyme activities related to cyanophycin metabolism in heterocysts and vegetative cells of Anabaena spp. J. Gen. Microbiol. 1981a;125:17–23. [Google Scholar]
- Gupta M. Carr NG. Enzymology of arginine metabolism in heterocyst-forming cyanobacteria. FEMS Microbiol. Lett. 1981b;12:179–181. [Google Scholar]
- Haselkorn R. Heterocyst differentiation and nitrogen fixation in cyanobacteria. In: Elmerich C, Newton WE, editors. Associative and endophytic nitrogen-fixing bacteria and cyanobacterial associations. The Netherlands: Springer; 2007. pp. 233–255. [Google Scholar]
- Hejazi M, Piotukh K, Mattow J, Deutzmann R, Volkmer-Engert R. Lockau W. Isoaspartyl dipeptidase activity of plant-type asparaginases. Biochem. J. 2002;364:129–136. doi: 10.1042/bj3640129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A. Flores E. Transport of basic amino acids by the dinitrogen-fixing cyanobacterium Anabaena PCC 7120. J. Biol. Chem. 1990;265:3931–3935. [PubMed] [Google Scholar]
- Herrero A, Picossi S. Flores E. Gene expression during heterocysts differentiation. Adv. Bot. Res. 2013;65:281–329. [Google Scholar]
- Jüttner F. 14C-labeled metabolites in heterocysts and vegetative cells of Anabaena cylindrica filaments and their presumptive function as transport vehicles of organic carbon and nitrogen. J. Bacteriol. 1983;155:628–633. doi: 10.1128/jb.155.2.628-633.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, et al. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8:205–213. doi: 10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- Ke S. Haselkorn R. The Sakaguchi reaction product quenches phycobilisome fluorescence, allowing determination of the arginine concentration in cells of Anabaena strain PCC 7120. J. Bacteriol. 2013;195:25–28. doi: 10.1128/JB.01512-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara S, Oda S, Kato K, Kim HG, Koyanagi T, Kumagai H, et al. A novel putrescine utilization pathway involves gammaglutamylated intermediates of Escherichica coli K-12. J. Biol. Chem. 2005;280:4602–4608. doi: 10.1074/jbc.M411114200. [DOI] [PubMed] [Google Scholar]
- Lang NJ, Simon RD. Wolk CP. Correspondence of cyanophycin granules with structured granules in Anabaena cylindrica. Arch. Microbiol. 1972;83:313–320. [Google Scholar]
- López-Igual R, Flores E. Herrero A. Inactivation of a heterocyst-specific invertase indicates a principal role of sucrose catabolism in the heterocysts of Anabaena sp. J. Bacteriol. 2010;192:5526–5533. doi: 10.1128/JB.00776-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Igual R, Lechno-Youssef S, Fan Q, Herrero A, Flores E. Wolk CP. A major facilitator superfamily protein, HepP, is involved in formation of heterocyst envelope polysaccharide in the cyanobacterium Anabaena sp. strain PCC7120. J. Bacteriol. 2012;194:4677–4687. doi: 10.1128/JB.00489-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinney G. Absorption of light by chlorophyll solutions. J. Biol. Chem. 1941;140:315–322. [Google Scholar]
- Markwell MAK, Hass SM, Bieber LL. Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Martín-Figueroa E, Navarro F. Florencio FJ. The GS-GOGAT pathway is not operative in the heterocysts. Cloning and expression of glsF gene from cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 2000;476:282–286. doi: 10.1016/s0014-5793(00)01722-1. [DOI] [PubMed] [Google Scholar]
- Merrit MV, Sid SS, Mesh L. Allen MM. Variations in the amino acid composition of cyanophycin in the cyanobacterium Synechocystis sp. PCC 6803 as a function of growth conditions. Arch. Microbiol. 1994;162:158–166. doi: 10.1007/BF00314469. [DOI] [PubMed] [Google Scholar]
- Messineo L. Modification of the Sakaguchi reaction: spectrophotometric determination of arginine in proteins without previous hydrolysis. Arch. Biochem. Biophys. 1966;117:534–540. [Google Scholar]
- Montesinos ML, Herrero A. Flores E. Amino acid transport systems required for diazotrophic growth in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1995;177:3150–3157. doi: 10.1128/jb.177.11.3150-3157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenchhoff J, Siddiqui KS, Poljak A, Raftery MJ, Barrow KD. Neilan BA. A novel prokaryotic L-arginine:glycine amidinotransferase is involved in cylindrospermopsin biosynthesis. FEBS J. 2010;277:3844–3860. doi: 10.1111/j.1742-4658.2010.07788.x. [DOI] [PubMed] [Google Scholar]
- Padan E, Raboy B. Shilo M. Endogenous dark respiration of the blue-green alga, Plectonema boryanum. J. Bacteriol. 1971;106:45–50. doi: 10.1128/jb.106.1.45-50.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernil R, Herrero A. Flores E. Catabolic function of compartmentalized alanine dehydrogenase in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2010;192:5165–5172. doi: 10.1128/JB.00603-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picossi S, Valladares A, Flores E. Herrero A. Nitrogen-regulated genes for the metabolism of cyanophycin, a bacterial nitrogen reserve polymer: expression and mutational analysis of two cyanophycin synthetase and cyanophycinase gene clusters in heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. J. Biol. Chem. 2004;279:11582–11592. doi: 10.1074/jbc.M311518200. [DOI] [PubMed] [Google Scholar]
- Quintero MJ, Muro-Pastor AM, Herrero A. Flores E. Arginine catabolism in the cyanobacterium Synechocystis sp. strain PCC 6803 involves the urea cycle and arginase pathway. J. Bacteriol. 2000;182:1008–1015. doi: 10.1128/jb.182.4.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raksajit W, Mäenpää P. Incharoensakdi A. Putrescine transport in a cyanobacterium Synechocystis sp. PCC 6803. J. Biochem. Mol. Biol. 2006;39:394–399. doi: 10.5483/bmbrep.2006.39.4.394. [DOI] [PubMed] [Google Scholar]
- Richter R, Hejazi M, Kraft R, Ziegler K. Lockau W. Cyanophycinase, a peptidase degrading the cyanobacterial reserve material multi-L-arginyl-poly-L-aspartic acid (cyanophycin): molecular cloning of the gene of Synechocystis sp. PCC 6803, expression in Escherichia coli, and biochemical characterization of the purified enzyme. Eur. J. Biochem. 1999;263:163–169. doi: 10.1046/j.1432-1327.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M. Stanier RY. Generic assignments, strain histories and properties of pure cultures of cianobacteria. J. Gen. Microbiol. 1979;111:1–61. [Google Scholar]
- Schilling N. Ehrnsperger K. Cellular differentiation of sucrose metabolism in Anabaena variabilis. Z. Naturforsch. 1985;40c:776–779. [Google Scholar]
- Schriek S, Rückert C, Staiger D, Pistorius EK. Michel KP. Bioinformatic evaluation of L-arginine catabolic pathways in 24 cyanobacteria and transcriptional analysis of genes encoding enzymes of L-arginine catabolism in the cyanobacterium Synechocystis sp. PCC 6803. BMC Genomics. 2007;8:437–465. doi: 10.1186/1471-2164-8-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska A, Danchin A. Risler JL. Phylogeny of related functions: the case of polyamine biosynthetic enzymes. Microbiology. 2000;146:1815–1828. doi: 10.1099/00221287-146-8-1815. [DOI] [PubMed] [Google Scholar]
- Sherman DM, Tucker D. Sherman LA. Heterocyst development and localization of cyanophycin in N2-fixing cultures of Anabaena sp. PCC 7120 (Cyanobacteria) J. Phycol. 2000;36:932–941. [Google Scholar]
- Thomas J, Meeks JC, Wolk CP, Shaffer PW, Austin SM. Chien W-S. Formation of glutamine from [13N]ammonia, [13N]dinitrogen and [14C] glutamate by heterocysts isolated from Anabaena cylindrica. J. Bacteriol. 1977;129:1545–1555. doi: 10.1128/jb.129.3.1545-1555.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares A, Rodríguez V, Camargo S, Martínez-Noël GMA, Herrero A. Luque I. Specific role of the cyanobacterial PipX factor in the heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 2011;193:1172–1182. doi: 10.1128/JB.01202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas WA, Nishi CN, Giarrocco LE. Salerno GL. Differential roles of alkaline/neutral invertases in Nostoc sp. PCC 7120: Inv-B isoform is essential for diazotrophic growth. Planta. 2011;233:153–162. doi: 10.1007/s00425-010-1288-5. [DOI] [PubMed] [Google Scholar]
- Weathers PJ, Chee HL. Allen MM. Arginine catabolism in Aphanocapsa 6308. Arch. Microbiol. 1978;118:1–6. doi: 10.1007/BF00406066. [DOI] [PubMed] [Google Scholar]
- Wegener KM, Singh AK, Jacobs JM, Elvitigala T, Welsh EA, Keren N, et al. Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol. Cell. Proteomics. 2010;9:2678–2689. doi: 10.1074/mcp.M110.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk CP, Ernst A. Elhai J. Heterocyst metabolism and development. In: Bryant DA, editor; The molecular biology of cyanobacteria. Dordrecht: Kluwer Academic Publishers; 1994. pp. 769–823. [Google Scholar]
- Yoon HS. Golden JW. Heterocyst pattern formation controlled by a diffusible peptide. Science. 1998;282:935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- Ziegler K, Diener A, Herpin C, Richter R, Deutzmann R. Lockau W. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-L-arginyl-poly-L-aspartate (cyanophycin) Eur. J. Biochem. 1998;254:154–159. doi: 10.1046/j.1432-1327.1998.2540154.x. [DOI] [PubMed] [Google Scholar]
- Ziegler K, Stephan DP, Pistorius EK, Ruppel HG. Lockau W. A mutant of the cyanobacterium Anabaena variabilis ATCC 29413 lacking cyanophycin synthetase: growth properties and ultrastructural aspects. FEMS Microbiol. Lett. 2001;196:13–18. doi: 10.1111/j.1574-6968.2001.tb10533.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction and verification of mutant CSMI11.
Microscopic observation of filaments of mutant strain CSMI11.
Catabolism of L-[14C]arginine in filaments of Anabaena wild type and strain CSMI11 incubated in medium lacking combined nitrogen.
Growth tests of Anabaena wild type and strain CSMI11 in solid media supplemented with putrescine.
Construction, verification and growth test of strain CSMI21.
Cyanobacterial strains and plasmids used in this work.
Oligodeoxynucleotide primers used in this work.


