Significance
Drug resistance is a major health problem, especially in quickly evolving disease targets including HIV-1 protease. Treatment regimens including HIV-1 protease inhibitors select for viral variants carrying mutations both in the protease and the substrates to confer drug resistance. We report the molecular mechanisms of this protease–substrate coevolution based on complex crystal structures of protease–substrate variants, complemented with molecular dynamics simulations. Specific interactions with I50V/A71V protease are observed to be lost or formed in response to coevolution mutations in the p1-p6 substrate cleavage site. Our structural analysis provides insights into how coevolution in HIV-1 may contribute to thwarting the effectiveness of available drug regimens.
Keywords: HIV-1 protease, drug resistance, coevolution, crystallography, active site dynamics
Abstract
Drug resistance mutations in response to HIV-1 protease inhibitors are selected not only in the drug target but elsewhere in the viral genome, especially at the protease cleavage sites in the precursor protein Gag. To understand the molecular basis of this protease–substrate coevolution, we solved the crystal structures of drug resistant I50V/A71V HIV-1 protease with p1-p6 substrates bearing coevolved mutations. Analyses of the protease–substrate interactions reveal that compensatory coevolved mutations in the substrate do not restore interactions lost due to protease mutations, but instead establish other interactions that are not restricted to the site of mutation. Mutation of a substrate residue has distal effects on other residues’ interactions as well, including through the induction of a conformational change in the protease. Additionally, molecular dynamics simulations suggest that restoration of active site dynamics is an additional constraint in the selection of coevolved mutations. Hence, protease–substrate coevolution permits mutational, structural, and dynamic changes via molecular mechanisms that involve distal effects contributing to drug resistance.
Resistant pathogens evolve under the selective pressure of drug therapies, commonly by acquiring mutations in the drug target (1–4). Most of these mutations cluster around the drug-binding site and alter key interactions between the drug and its target. Strikingly, mutations in other off-target proteins have also been reported to contribute to drug resistance (5–8) where the mechanism of resistance is not as straightforward to rationalize. In the case of HIV-1, mutations in the target protease gene confer resistance to protease inhibitors (PIs) and correlate with emergence of mutations elsewhere in Gag. There are currently nine protease inhibitors (PIs) that are US Food and Drug Administration (FDA) approved for clinical use including in highly active antiretroviral therapy (HAART) (9), and all are competitive inhibitors binding at the active site.
HIV-1 protease is a key antiviral drug target due to its essential function of processing Gag and Gag-Pol polyproteins in viral maturation (10–12). Under the selective pressure of PI-including therapy regimens, viral variants carrying mutations in the protease gene impair the inhibitor efficacy. Although the PIs become weaker binders of these resistant protease variants, the substrates are still hydrolyzed (13, 14), skewing the balance between inhibitor binding and substrate processing in favor of the latter. Earlier work from our group revealed the molecular determinants of this fine balance and formulated the substrate envelope hypothesis to effectively explain the molecular mechanism of resistance due to primary active site mutations (15). Among primary protease mutations, I50V is commonly observed in patients failing therapy with the PIs amprenavir (APV) and darunavir (DRV) (16–18). Residue 50 is located at the flap tip of the flexible loop (50s loop) that controls the access of substrates and competitive inhibitors to the protease active site. In addition to conferring resistance to PIs, the I50V mutation also impairs substrate processing (19). The loss of catalytic efficiency due to I50V is compensated by secondary mutations, in particular A71V (20), which is observed in more than 50% of patient sequences bearing I50V (14).
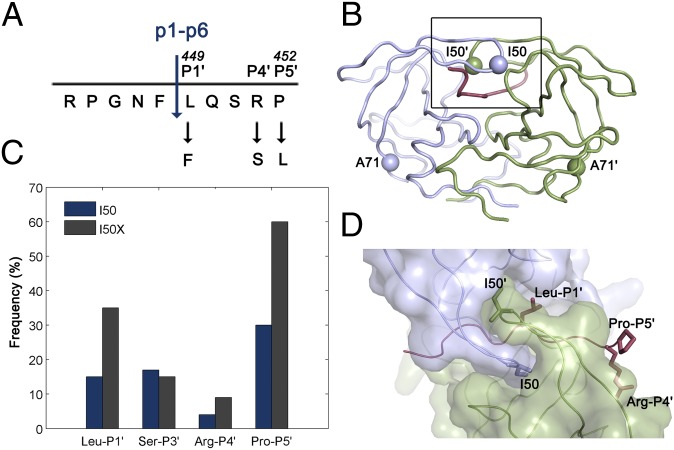
Several mutations in Gag both within cleavage sites and elsewhere coevolve with primary protease mutations contributing to viral fitness and possibly drug resistance (5, 7, 21–25). Particularly, mutations in the p1-p6 cleavage site are statistically associated with I50V protease mutation in the viral sequences retrieved from patients (Fig. 1) (26). The Gag L449F mutation rescues the protease activity by 10-fold, whereas P453L, despite being distal from the catalytic site, causes a 23-fold enhancement (19). However, the molecular basis for the selection advantage of these correlated mutations and the mechanism by which the compensatory mutations restore substrate recognition in drug resistance is not clear. In this study, we report the structural basis for the coevolution of I50V/A71V protease with the p1-p6 substrate. Through a series of cocrystal structures, the Gag mutations L449F and P453L were shown to enhance van der Waals (vdW) interactions between the substrate and mutant protease, whereas R452S results in an additional hydrogen bond. Unexpectedly, the P453L substrate mutation causes a conformational change in the protease flap loop, revealing the molecular mechanism by which this distal substrate mutation is able to enhance substrate–protease interactions. In addition, molecular dynamics simulations suggest that coevolution restores the dynamics at the active site, a key aspect of substrate recognition and turnover that is largely uncharacterized.
Fig. 1.
HIV-1 protease and p1-p6 cleavage site coevolution with I50V primary drug resistance mutation. (A) p1-p6 cleavage site sequence and the most common coevolution mutations at P1′, P4′, and P5′ sites. (B) Residues 50 and 71 are indicated as spheres on the homodimeric HIV-1 protease structure. (C) Frequency of mutations in the p1-p6 cleavage site without (dark blue) and with (gray) any mutations at position 50 of the protease. The difference is statistically significant for LP1′, RP4′, and PP5′. Data from ref. 26. (D) Side chains of the substrate residues LP1′, RP4′, and PP5′ and the protease residue I50 are shown as sticks. Monomers of HIV-1 protease are in light purple and green, and the substrate is red in B and D.
Results
To understand how HIV-1 protease–substrate coevolution alters binding interactions, a series of cocrystal structures of catalytically inactive (D25N) WT and I50V/A71V protease with p1-p6 substrate variant peptides (WT, L449F, R452S, and P453L) was determined. All structures were solved to resolution 1.50–2.14 Å (Table S1). In addition to an overall structural comparison, the alterations in coevolved substrates’ fit within the substrate envelope and details of molecular interactions between the protease and substrate were analyzed. Whereas the D25N mutation to prevent substrate cleavage may have minor effects on the structures (27), this change has been incorporated in all complexes; therefore, observed changes are expected to be the direct result of the coevolution mutations. Finally, molecular dynamics (MD) simulations initiated from these crystal structures were performed to reveal any dynamic changes in coevolved complexes relative to WT.
The Overall Structure and Substrate Envelope Is Conserved in Coevolved Complexes.
The overall backbone conformation of substrate–protease complexes is conserved in all coevolved structures. When the structures are superposed onto the WTWT complex (based on the structurally conserved regions, residues 24–26 and 85–90) (28), the root mean square deviation (RMSD) of Cα atoms is within 0.48 Å. Minor structural changes in some coevolved structures are located mainly at crystal contact surfaces (Fig. S1). One notable exception is in the I50V/A71VPP5′L structure, where a peptide bond flips to change the flap loop conformation relative to the WT structure in one monomer. Similar to the protease, all substrate residues from P4 to P4′ overlap well, with the exception of P4′ arginine having an altered orientation in the WTWT complex (Fig. S2). The distal P5 and P5′ residues are more flexible; P5 is often disordered, whereas P5′ has altered conformations in both LP1′F and PP5′L substrates as detailed below.
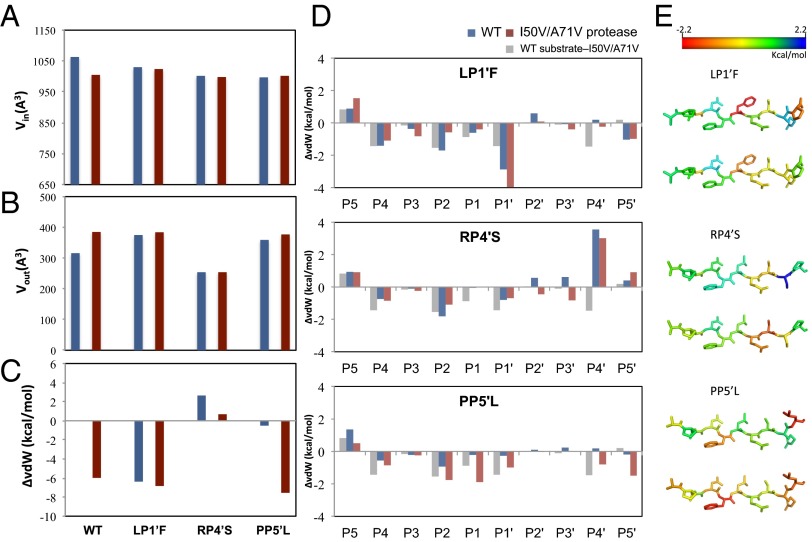
The protease and substrate mutations were evaluated for their effect on the substrate’s fit to the substrate envelope, which is key to recognition by the protease (Fig. 2 A and B). The substrate volume within the envelope, Vin, is the largest in the WTWT complex, indicating that the WT substrate fills the substrate envelope better than the other substrate variants. The I50V/A71V protease mutations worsen the fit of the WT substrate within the substrate envelope, resulting in a 53-Å3 reduction in Vin. The LP1′F substrate partially restores this loss in Vin and the coevolved substrate better fills the substrate envelope by 25 Å3. Overall, the coevolved substrates maintain a comparable fit within the substrate envelope regardless of whether the protease carries the I50V/A71V mutations or not, supporting that the substrate envelope is the recognition motif, and coevolved mutations at the cleavage site do not drastically alter the fit within the substrate envelope.
Fig. 2.
HIV-1 protease–p1-p6 substrate interactions at the active site. The fit of HIV-1 protease substrates within the substrate envelope evaluated by (A) volume within the envelope (Vin), and (B) volume protruding outside the envelope (Vout). (C) The change in total vdW contacts between the substrate and protease relative to the WT complex. Negative values indicate enhanced interactions. (D) The change in vdW contacts of substrate residues relative to the WT complex. (E) The difference between vdW contact changes depicted on the substrate structure. (Upper) For the changes in the coevolved complex relative to I50V/A71VWT (difference between the red and gray bars in D). (Lower) Relative to WT protease with the corresponding mutant substrate (red and blue bars in D).
Substrate Mutations Have Distal Effects and Enhance Packing at the Active Site.
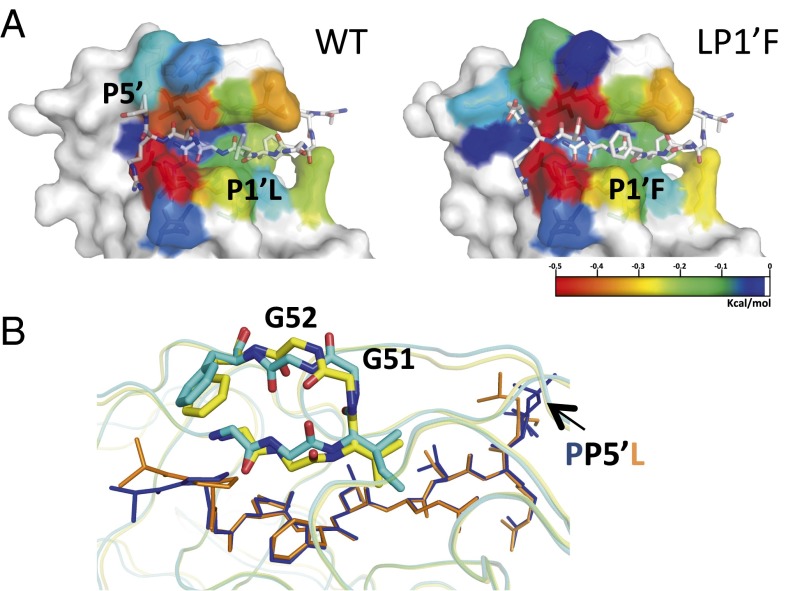
To investigate the alterations in substrate packing at the active site, the vdW interactions were quantified between the protease and substrate. The changes in substrate packing relative to WTWT structure are displayed in Fig. 2C, where negative values indicate enhanced packing. The LP1′F substrate has more vdW contacts with I50V/A71V protease compared with those in either I50V/A71VWT or WTLP1′F. Similarly, for the PP5′L substrate, the coevolved I50V/A71VPP5′L complex makes more vdW contacts than either the I50V/A71VWT or WTPP5′L complexes. Hence, the coevolution of HIV-1 protease and p1-p6 substrate may rescue the loss of binding interactions caused by mutations on either protease or substrate alone. The enhanced interactions of I50V/A71VLP1′F was partially due to the LP1′F mutation (Fig. 2D), which has more interactions with residues that surround the S1′ pocket Arg8, Leu23, Pro81, and Val82 compared with I50V/A71VWT and more interaction with Val82 compared with WTLP1’F (Fig. S3). In addition, the LP1′F mutation causes a distal change at the substrate P5′ proline (Figs. 2 D and E and 3A), which is in an alternative position in the WT complex (Fig. S2). This change increases the P5′ proline’s vdW contacts, specifically with Lys45′ and Met46′ instead of Phe53′. Hence, the effect of LP1′F mutation is not localized solely to the site of mutation. This alteration of the structure and vdW contacts of residues at a distal position illustrates the adaptability and interdependency of interactions when HIV-1 protease recognizes substrates.
Fig. 3.
Distal effects of substrate mutations on I50V/A71V protease interactions. (A) The vdW contacts of residues in HIV-1 protease–substrate cocrystal structures colored blue to red for increasing contacts. The substrate mutation at P1′ position enhances contacts at P5′. (B) The distal substrate mutation PP5′L causes a conformational change in the protease flap and alters substrate–protease interactions. The protease flaps are in cyan and yellow in complex structures with WT (navy blue) and P5′L (orange) substrates, respectively.
The RP4′S substrate has more overall vdW contacts in the coevolved I50V/A71VRP4′S complex relative to WTRP4′S but less than the WT substrate in I50V/A71VWT. Because P4′ serine is a smaller residue compared with arginine, the mutated serine makes less vdW contacts when bound to either WT or I50V/A71V protease compared with arginine with the corresponding protease (Fig. 2C). However, the mutation at the P4′ residue actually influences the interactions at other positions: P3′ Ser and P2′ Gln have enhanced vdW contacts in I50V/A71VRP4′S compared with either I50V/A71VWT or WTRP4′S. Specifically, P3′ Ser makes more contacts with Asp29′ compared with I50V/A71VWT, whereas P2′Gln–Asp30′ and P3′Ser–Arg8/Ile47′ interactions are enhanced compared with WTRP4′S (Fig. S3). As with the LP1′F mutation, the RP4′S mutation impacts the interactions of other substrate residues with the protease.
The PP5′L mutation increases the P5′ residue’s interactions with the I50V/A71V protease as the leucine packs closer to the 50s loop in one monomer while influencing the residues closer in the active site, P2 Asn and P1 Leu, to also form more extensive contacts. In addition, the coevolved structure makes more contacts at the P1′ residue compared with WTPP5′L. Interestingly, the major structural change due to this substrate mutation is observed within the protease (Fig. 3B). The peptide bond between Gly51 and Gly52 in the I50V/A71VPP5′L structure is flipped compared with the other structures, and this flipped peptide bond pushes the 50s loop toward the substrate, causing increased vdW contacts between protease residues Gly48, Gly49, Ile50, Phe53, and the substrate. Hence, the coevolved site not only impacts its own fit within protease active site but also alters the interactions of distal residues in the substrate by stabilizing alternative conformations of the protease.
In conclusion, the detailed analysis of vdW contacts between the protease and substrates shows interdependent distal effects in binding interactions where the alterations are not localized at the mutated residue itself but also occur at other residues. These distal alterations are caused by structural changes in the protease, the substrate, or both.
The RP4′S Substrate Has Fewer Packing Interactions but an Additional Hydrogen Bond.
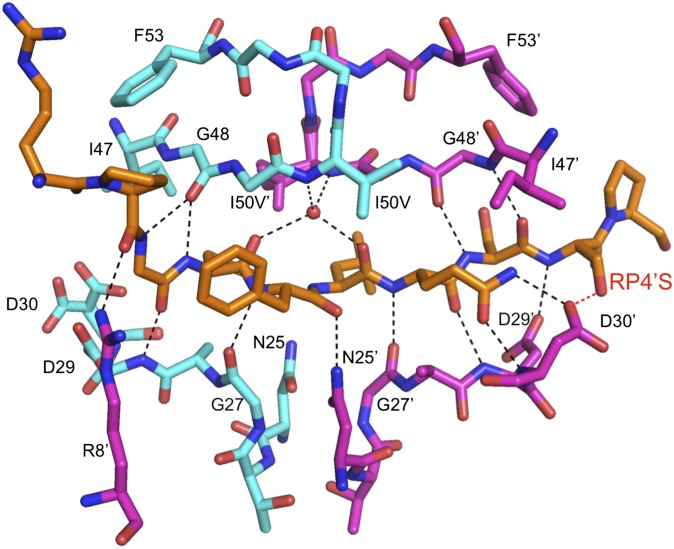
The intramolecular hydrogen bonds between the protease and the substrates are conserved across all complexes, with two exceptions (Fig. 4 and Table S2). First, the P4′ arginine in the WTWT structure orients in the opposite direction compared with the other complexes, making a hydrogen bond with the side chain of Asp30′ instead of Asp29′. Second, the RP4′S substrate forms an additional hydrogen bond with both WT and I50V/A71V protease through the P4′ serine side chain (Fig. 4). This extra hydrogen bond may compensate for the loss of vdW contacts due to the smaller size of serine in these two complexes.
Fig. 4.
The hydrogen bonds between p1-p6 substrate (orange) and HIV-1 protease (cyan and magenta monomers), including those mediated by a conserved water molecule (red sphere). Bonds shared in all substrate variants are in black, whereas the additional hydrogen bond formed by RP4'S variant is indicated in red.
Active Site Dynamics Is Restored by Coevolution.
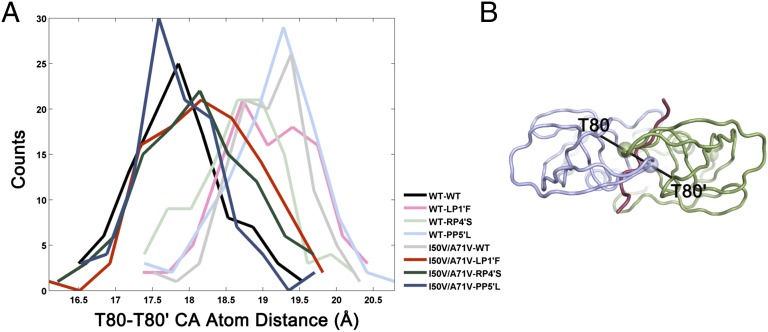
Resistance mutations, in addition to altering the molecular interactions, affect the conformational dynamics at the active site. Specifically, the distance between the two 80s loops, which reflects the overall size of the protease active site, varies between protease–substrate complexes during the MD simulations (Fig. 5). In crystal structures, the 80–80′ distance is similar in all of the structures and varies between 17.1 and 18.0 Å. In the dynamic conformational ensemble of the WTWT structure, the distance between these two loops is around 17.5 Å and expands to 19–19.5 Å with mutations in either the protease or substrate. Strikingly, in all three cases, coevolution brings this distance back to 17.5–18.0 Å, which is similar to the WT interloop distance (Fig. 5). Hence, mutation of either the protease or the substrate alone disturbs the dynamics of the protease active site, whereas coevolution of both restores the active site dynamics and possibly the protease activity.
Fig. 5.
The distance (in angstroms) distribution between T80 and T80′ across the active site during MD simulations of substrate–protease complexes. Counts refer to the number of MD snapshots for a given distance. (A) Mutations in only protease or only substrate increase the distance sampled (lighter shades), whereas coevolution of both together (darker shades) brings the distance back to that in the WT–WT complex (black). (B) The T80–T80′ distance across the active site depicted on the protease with a view from the top of the flaps. The colors are the same as in Fig. 1.
Discussion
The HIV-1 I50V/A71V protease is commonly observed in patients failing therapy with APV and DRV, and substrate mutations in Gag cleavage sites coevolve with these primary protease mutations to contribute to inhibitor resistance. Gag L449F mutation rescues the protease activity by 10-fold, whereas P453L, although located distal from the catalytic site, causes a 23-fold enhancement (19). The mutated substrates are cleaved more efficiently than the WT substrate by the I50V/A71V protease (19). Interestingly, the WT protease also processes the WT substrate less efficiently than the mutant substrates. This suboptimal cleavage efficiency at the p1-p6 site should be important for temporal regulation of Gag processing preventing premature viral maturation (11, 29). Under drug pressure, the resistance mutations I50V/A71V are populated, and the impaired protease activity on the WT Gag may interfere with the ordered processing of Gag. Coevolution of substrates possibly restores proper Gag processing by getting more efficiently cleaved by the protease.
The molecular basis of such protease–substrate coevolution was investigated in this study by solving the crystal structures of the complexes of WT and I50V/A71V protease with the p1-p6 substrate variants combined with molecular dynamics simulations. Although all complexes have similar overall backbone structures, vdW contacts and hydrogen bonds between the protease and substrate are altered. In fact, although the overall vdW interactions with the WT substrate increase in the I50V/A71V complex, a key intramolecular hydrogen bond is lost, and the active site dynamics of this complex are disturbed. The coevolved complex structures display restored active site dynamics and enhanced overall substrate interactions, due to either more vdW contacts or hydrogen bonds, compared with complexes with mutations in either the protease or the substrate alone.
Coevolving mutations in the substrate are not selected to restore the specific interactions lost due to drug resistance mutations but instead enhance substrate–protease interactions through a variety of molecular mechanisms. P1′ and P5′ mutations enhance substrate packing at the active site, whereas P4′ contributed an additional hydrogen bond with the protease. A similar compensation of interactions is observed in substrate coevolution with nelfinavir resistant D30N/N88D protease, where lost interactions were compensated for by new interactions particularly at the site of substrate coevolution mutation (30, 31).
In I50V/A71V protease coevolution, the effects of substrate mutations are not local, but propagate to distal parts of both the substrate and the protease. The mutated P4′ serine affects the interactions at other positions, particularly at P3′ Ser and P2′ Gln, which has enhanced vdW contacts in I50V/A71VRP4′S compared with I50V/A71VWT and WTRP4′S. Substrate positions P1′ and P5′ mutually influence each other’s interactions with the protease. Mutation at either residue results in enhancement of vdW contacts at both sites: In the presence of the LP1′F mutation the P5′ residue and in the presence of the PP5′L mutation, the P1′ residue's packing is altered. Apart from these synergistic effects within the substrate, the P5′ mutation in the substrate stabilized an unexpected structural change within the protease. The protease structure accommodated this mutation by flipping the peptide bond between Gly51 and Gly52 in one of the flaps. This backbone flip brings the flap closer to the substrate and increases vdW contacts at the P2, P1, and P1′ positions. Hence, the HIV-1 protease adopts a conformational change to favor the substrate’s binding in the coevolved complex. This ability of mutations to have distal effects explains why a coevolution mutation at the P5′ position, which is away from the core region of the substrate at the active site, may be selected and how this distal mutation is able to alter substrate–protease interactions.
HIV-1 protease is a highly dynamic protein, and conformational dynamics, especially around the active site, is crucial to substrate binding and enzymatic activity (32–36). Although crystal structures provide key insights, alteration of dynamic behavior, not captured by static structures, is emerging as an additional contribution to mechanisms of drug resistance (37, 38). We found that drug resistance mutations in the protease or in the native substrate disturbed the active site dynamics, which was restored in all coevolved complexes bearing complementary mutations in both the protease and the substrate. These results suggest that, in addition to the specific 3D shape adopted and shared by all substrates when bound to the HIV-1 protease, as defined by the substrate envelope, a conserved dynamic behavior around the active site may be an additional substrate recognition and selection constraint. This dynamic constraint may contribute to the selection of these specific substrate coevolution mutations in response to the disturbed dynamics in mutated drug resistant protease.
Previously, we showed that the I50V/A71V protease has decreased vdW interactions with the protease inhibitors APV and DRV compared with WT (0.61 and 1.98 kcal/mol, respectively), mainly due to the loss of a methyl group interacting with the sulfonyl moiety in APV/DRV (39). The coevolved I50V/A71VLP1′F and I50V/A71VPP5′L structures have more vdW contacts, and I50V/A71VRP4′S has more hydrogen bonds compared with WTWT complex. Unlike substrates, the inhibitors cannot adapt to conformational changes in the drug-resistant protease, such as the peptide bond flip in the flap of I50V/A71VPP5′L protease. Hence, the structural adaptability of the protease–substrate system allows drug resistance to evolve by selecting mutations in the protease that decrease inhibitor affinity and additional compensatory mutations in the substrate to enhance any inadvertently lost substrate interactions through various molecular mechanisms, including propagating distal effects.
Coevolution causes distal changes both in the substrate and the protease, and adaptability of the complex permits mutational, structural, and dynamic plasticity to confer drug resistance. Therefore, the resistance mechanism is an interdependent process whereby multiple residues act in concert on both sides. The molecular rationale, reported here, for the distal effects of mutations in the nontarget polyprotein under HIV-1 PI treatment should provide insights into allosteric events in a wider range of coevolving systems where function is maintained by complex interdependent protein interactions.
Materials and Methods
Nomenclature.
The HIV-1 protease (WT or I50V/A71V) complexes with different p1-p6 substrate variants [WT, L449F (LP1′F), R452S (RP4′S), and P453L (PP5′L)] are distinguished with subscripts. For example, the WT protease in complex with LP1′F p1-p6 substrate is denoted by WTLP1′F and I50V/A71V protease in complex with the WT p1-p6 substrate is denoted by I50V/A71VWT. The HIV-1 protease functions as a homodimer, and residues in monomer A are simply indicated by residue number, whereas residues in monomer B are marked with the residue number with a prime. For example, arginine 8 in monomer A is Arg8, and valine 50 in monomer B is Val50′.
Substrate Peptides.
Substrate peptides of the p1-p6 processing site within the Gag polyprotein (amino acids 444–453) and its variants were purchased from Quality Controlled Biochemicals. The substrate sequences of WT and coevolved substrates are (i) p1-p6WT–RPGNFLQSRP, (ii) p1-p6LP1′F–RPGNFFQSRP, (iii) p1-p6RP4′S–RPGNFLQSSP, and (iv) p1-p6PP5′L–RPGNFLQSRL.
Protease Gene Construction.
The synthetic protease gene was constructed using codon optimization for protein expression in Escherichia coli, and the Q7K mutation was introduced to prevent autoproteolysis (40). For protease–substrate cocrystallization purposes, the D25N mutation was introduced to prevent substrate cleavage; this mutation has a negligible impact on protease structure (27). I50V/A71V protease mutations were introduced sequentially by using the QuikChange site-directed mutagenesis kit (Stratagene).
Protein Expression, Purification, and Crystallization.
The gene encoding the HIV protease was subcloned into the heat-inducible pXC35 expression vector (ATCC) and transformed into E. coli TAP-106 cells. Protein expression and purification were performed as previously described (41). Protease purified from size exclusion column (equilibrated with gel filtration buffer containing 0.05 M sodium acetate at pH 5.5, 5% (vol/vol) ethylene glycol, 10% (vol/vol) glycerol, and 5 mM DTT) was concentrated to 2 mg/mL using an Amicon Ultra-15 10-kDa device (Millipore) for crystallization. The concentrated samples were incubated with 10 molar excess of substrates overnight at 4 °C. The concentrated protein solution was then mixed with either precipitant solution [126 mM sodium phosphate buffer pH 6.2, 63 mM sodium citrate, 20–32% (wt/vol) ammonium sulfate; or 0.1 M citrate phosphate pH 5.5, 0.5–3.0 M ammonium sulfate] at a 1:1 ratio in 24-well VDX hanging-drop trays (Hampton Research) at room temperature. Diffraction quality crystals were obtained within 1 wk.
Data Collection and Structure Solution.
Diffraction quality crystals were flash-frozen in liquid nitrogen for storage. A constant cryostream was applied when mounting the crystal, and X-ray diffraction data were collected at the Advanced Photon Source LS-CAT 21-ID-F or at our in-house Rigaku_Saturn 944 X-ray system. The substrate complexes’ diffraction intensities were indexed, integrated, and scaled using the program HKL2000 (42). The number of molecules in the asymmetric unit was determined by the Matthews coefficient calculation. The structure solutions were generated using simple isomorphous molecular replacement with PHASER (43). WT protease–DRV cocrystal structure was used as the starting model [Protein Data Bank (PDB) ID code 1T3R]. Initial refinement was carried out in the absence of modeled substrate, which was subsequently built in during later stages of refinement. On obtaining the correct molecular replacement solutions, ARP/wARP or Phenix (44) was applied to improve the phases by building solvent molecules (45). Crystallographic refinement was carried out using the CCP4 program suite or PHENIX with iterative rounds of translation–libration–screw and restrained refinement until convergence was achieved (46). MolProbity (47) was applied to evaluate the final structures before deposition in the PDB. Five percent of the data were reserved for the free R-value calculation to limit the possibility of model bias throughout the refinement process (48). Interactive model building and electron density viewing were carried out with COOT (49).
Structural Analysis.
Hydrogen bonds were determined using Maestro (Suite 2012: Maestro, version 9.3, Schrödinger). A hydrogen bond was defined by a distance between the donor and acceptor of less than 3.5 Å and a donor–hydrogen–acceptor angle of greater than 120°.
The vdW contacts between the protease and substrate were estimated using a simplified Lennard-Jones potential V(r) = 4ε[(σ/r)12 – (σ/r)6], with the well depth (ε) and hard sphere diameter (σ) for each protease–substrate atom pair. V(r) for all protease–substrate atom pairs was computed within 6 Å, and when the distance between nonbonded pairs was less than ε, V(r) was considered to be equal to –ε. The rationale for this modification to the original 6–12 Lennard-Jones potential was previously described in detail (28). Using this simplified potential for each nonbonded pair, ΣV(r) was then computed for the protease–substrate complex.
The HIV-1 protease substrate envelope was defined using a 3D grid, and the fit of a substrate within this substrate envelope for a given cocrystal structure was evaluated using Vin and Vout (volumes of the substrate within and outside the substrate envelope, respectively), as previously described in detail (28). Only the P4 to P4′ residues of the substrates were modeled in the substrate envelope, because the substrate residues beyond these positions do not share a significant consensus volume.
Molecular Dynamics Simulations.
The crystal structures were prepared for simulations by keeping the crystallographic waters within 4.0 Å of any protease or substrate atom but removing the buffer salts from the coordinate file. The structures were further processed with the Protein Preparation Tool from Schrodinger by adding hydrogen atoms, building side chains with missing atoms, and determining the optimal protonation states for the ionizable side chains. The hydrogen bonding network of the initial structures was optimized by flipping the terminal chi angle of Asn, Gln, and His residues and sampling hydroxyl/thiol polar hydrogens with the exhaustive/water orientational sampling options. Before solvation, the structures were minimized in vacuum with restraints on heavy atoms using the Impact refinement module with the OPLS2005 force field until the RMSD reached 0.3 Å, allowing the hydrogens to be freely minimized while relaxing the strained bonds, angles, and potential clashes. The prepared systems were solvated in a truncated octahedron solvent box with the SPC water model extending 10 Å beyond the protein in all directions, using the System Builder utility. The overall charge was neutralized by adding the necessary number of counter ions (Na+ or Cl–).
Desmond was used in all simulations with the OPLS2005 force field. Each system was first relaxed using a protocol consisting of an initial minimization restraining the solute heavy atoms with a force constant of 1,000 kcal⋅mol−1⋅Å−2 for 10 steps with the steepest descent and with the limited-memory Broyden–Fletcher–Goldfarb–Shanno (LBFSG) method up to 2,000 total steps with a convergence criterion of 50.0 kcal⋅mol−1⋅Å−2. The system was further minimized by restraining only the backbone and allowing the free motion of the side chains. At this stage, the restraint on the backbone was gradually reduced from 1,000 to 1.0 kcal⋅mol−1⋅Å−2 in 5,000 steps (250 steepest descent plus 4,750 LBFSG) for each value of force constant (1,000, 500, 250, 100, 50, 10, and 1.0 kcal⋅mol−1⋅Å−2), and finally an unrestrained energy minimization was performed.
After energy minimization, each system was equilibrated by running a series of short MD steps. First, a 10-ps MD simulation at 10 K was performed with a restraint (50 kcal⋅mol−1⋅Å−2) on solute heavy atoms and using a Berendsen thermostat in the constant-temperature, constant-volume (NVT) ensemble. MD steps were integrated using a two time-step algorithm, with 1-fs steps for bonded and short-range interactions within the 9-Å cutoff and 3 fs for long-range electrostatic interactions, which were treated with the smooth particle-mesh Ewald (PME) method. Time steps were kept shorter at this first MD stage to reduce the numerical issues associated with large initial forces before the system equilibrates. This equilibration was followed by another restrained MD simulation for 10 ps at 10 K with a 2-fs inner and 6-fs outer time step in the constant-temperature, constant-pressure (NPT) ensemble. The temperature of the system was slowly increased from 10 to 300 K over 10 ps, retaining the restraint on the system, and 10-ps MD was performed without the harmonic restraints. Production MD simulations were carried out at 300 K and 1 bar for 20 ns using the NPT ensemble, a Nose-Hoover thermostat, and a Martyna-Tuckerman-Klein barostat. The long-range electrostatic interactions were computed using a smooth PME approximation with a cutoff radius of 9 Å for the transition between the particle–particle and particle–grid calculations, and vdW interactions were truncated at 9 Å. The coordinates and energies were recorded every 5 ps.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health Grant R01-GM65347.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4QJ9, 4QJA, 4QJ2, 4QJ6, 4QJ7, and 4QJ8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414063111/-/DCSupplemental.
References
- 1.Theuretzbacher U, Mouton JW. Update on antibacterial and antifungal drugs - can we master the resistance crisis? Curr Opin Pharmacol. 2011;11(5):429–432. doi: 10.1016/j.coph.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Jänne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov. 2009;8(9):709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 3.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105(6):2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali A, et al. Molecular basis for drug resistance in HIV-1 protease. Viruses. 2010;2(11):2509–2535. doi: 10.3390/v2112509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dam E, et al. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Path 5(3):e1000345.
- 6.Kern WV, Oethinger M, Jellen-Ritter AS, Levy SB. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2000;44(4):814–820. doi: 10.1128/aac.44.4.814-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolli M, Lastere S, Schiffer CA. Co-evolution of nelfinavir-resistant HIV-1 protease and the p1-p6 substrate. Virology. 2006;347(2):405–409. doi: 10.1016/j.virol.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 8.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: An update. Drugs. 2009;69(12):1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debouck C. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res Hum Retroviruses. 1992;8(2):153–164. doi: 10.1089/aid.1992.8.153. [DOI] [PubMed] [Google Scholar]
- 10.Chou KC, Tomasselli AG, Reardon IM, Heinrikson RL. Predicting human immunodeficiency virus protease cleavage sites in proteins by a discriminant function method. Proteins. 1996;24(1):51–72. doi: 10.1002/(SICI)1097-0134(199601)24:1<51::AID-PROT4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Pettit SC, Sheng N, Tritch R, Erickson-Viitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv Exp Med Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- 12.Sadler BM, Stein DS. Clinical pharmacology and pharmacokinetics of amprenavir. Ann Pharmacother. 2002;36(1):102–118. doi: 10.1345/aph.10423. [DOI] [PubMed] [Google Scholar]
- 13.Kantor R, et al. Evolution of primary protease inhibitor resistance mutations during protease inhibitor salvage therapy. Antimicrob Agents Chemother. 2002;46(4):1086–1092. doi: 10.1128/AAC.46.4.1086-1092.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee SY, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31(1):298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: Lessons from HIV-1 protease. Chem Biol. 2004;11(10):1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Partaledis JA, et al. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J Virol. 1995;69(9):5228–5235. doi: 10.1128/jvi.69.9.5228-5235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Marck H, et al. Unravelling the complex resistance pathways of darunavir using bioinformatics resistance determination (BIRD) Antivir Ther. 2007;12(5):S141. [Google Scholar]
- 18.Vermeiren H, et al. Virco Clinical Response Collaborative Team Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J Virol Methods. 2007;145(1):47–55. doi: 10.1016/j.jviromet.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Maguire MF, et al. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J Virol. 2002;76(15):7398–7406. doi: 10.1128/JVI.76.15.7398-7406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nijhuis M, et al. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS. 1999;13(17):2349–2359. doi: 10.1097/00002030-199912030-00006. [DOI] [PubMed] [Google Scholar]
- 21.Bally F, Martinez R, Peters S, Sudre P, Telenti A. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: Clinical significance and implications for resistance to protease inhibitors. AIDS Res Hum Retroviruses. 2000;16(13):1209–1213. doi: 10.1089/08892220050116970. [DOI] [PubMed] [Google Scholar]
- 22.Doyon L, et al. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70(6):3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fehér A, et al. Effect of sequence polymorphism and drug resistance on two HIV-1 Gag processing sites. Eur J Biochem. 2002;269(16):4114–4120. doi: 10.1046/j.1432-1033.2002.03105.x. [DOI] [PubMed] [Google Scholar]
- 24.Mammano F, Petit C, Clavel F. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: Phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J Virol. 1998;72(9):7632–7637. doi: 10.1128/jvi.72.9.7632-7637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YM, et al. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71(9):6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolli M, Stawiski E, Chappey C, Schiffer CA. Human immunodeficiency virus type 1 protease-correlated cleavage site mutations enhance inhibitor resistance. J Virol. 2009;83(21):11027–11042. doi: 10.1128/JVI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J Biol Chem. 2008;283(19):13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozen A, Haliloğlu T, Schiffer CA. Dynamics of preferential substrate recognition in HIV-1 protease: Redefining the substrate envelope. J Mol Biol. 2011;410(4):726–744. doi: 10.1016/j.jmb.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology. 2005;2:66. doi: 10.1186/1742-4690-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolli M, Ozen A, Kurt-Yilmaz N, Schiffer CA. HIV-1 protease-substrate coevolution in nelfinavir resistance. J Virol. 2014;88(13):7145–7154. doi: 10.1128/JVI.00266-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozen A, Haliloğlu T, Schiffer CA. HIV-1 Protease and Substrate Coevolution Validates the Substrate Envelope As the Substrate Recognition Pattern. J Chem Theory Comput. 2012;8(2):703–714. doi: 10.1021/ct200668a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foulkes-Murzycki JE, Scott WR, Schiffer CA. Hydrophobic sliding: A possible mechanism for drug resistance in human immunodeficiency virus type 1 protease. Structure. 2007;15(2):225–233. doi: 10.1016/j.str.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freedberg DI, et al. Rapid structural fluctuations of the free HIV protease flaps in solution: Relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11(2):221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishima R, Freedberg DI, Wang YX, Louis JM, Torchia DA. Flap opening and dimer-interface flexibility in the free and inhibitor-bound HIV protease, and their implications for function. Structure. 1999;7(9):1047–1055. doi: 10.1016/s0969-2126(99)80172-5. [DOI] [PubMed] [Google Scholar]
- 35.Mittal S, Cai Y, Nalam MN, Bolon DN, Schiffer CA. Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease. J Am Chem Soc. 2012;134(9):4163–4168. doi: 10.1021/ja2095766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perryman AL, Lin JH, McCammon JA. HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: Possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 2004;13(4):1108–1123. doi: 10.1110/ps.03468904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai Y, Yilmaz NK, Myint W, Ishima R, Schiffer CA. Differential Flap Dynamics in Wild-type and a Drug Resistant Variant of HIV-1 Protease Revealed by Molecular Dynamics and NMR Relaxation. J Chem Theory Comput. 2012;8(10):3452–3462. doi: 10.1021/ct300076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Vera IMS, et al. Elucidating a relationship between conformational sampling and drug resistance in HIV-1 protease. Biochemistry. 2013;52(19):3278–3288. doi: 10.1021/bi400109d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mittal S, et al. Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50. J Virol. 2013;87(8):4176–4184. doi: 10.1128/JVI.03486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosé JR, Salto R, Craik CS. Regulation of autoproteolysis of the HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J Biol Chem. 1993;268(16):11939–11945. [PubMed] [Google Scholar]
- 41.King NM, et al. Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease. Protein Sci. 2002;11(2):418–429. doi: 10.1110/ps.25502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol Macromolecular Crystallography Part A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP’s model-building algorithms. I. The main chain. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 6 Pt 2):968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 46.Bailey S. Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375-83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brünger AT. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355(6359):472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.