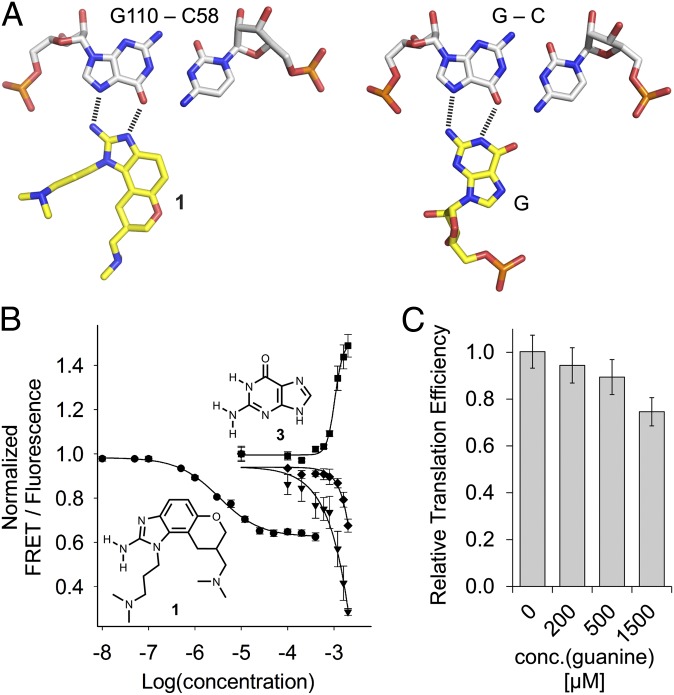
Fig. 2.
Ligand binding to the HCV IRES. (A) Docking of the benzimidazole 1 at the C58-G110 base pair in the crystal structure of the HCV subdomain IIa target complex in comparison with the geometry of a C-G○G triple. (B) Titrations of Cy3/Cy5-labeled HCV subdomain IIa RNA with benzimidazole 1 and guanine 3. Curves show normalized FRET signal for 1 (●) and guanine (▼), as well as the normalized fluorescence signals of the donor Cy3 (■) and acceptor Cy5 (♦) for guanine. Fitting of a single-site binding curve to the benzimidazole 1 FRET response gave an EC50 value for ligand binding of 3.4 ± 0.3 μM. Because the FRET signal did not reach saturation in the guanine titration, affinity was estimated by fitting to the Cy3 emission, which resulted in an EC50 value for ligand binding of 1,050 ± 69 μM (guanine). (C) Effect of guanine on IRES-driven translation, as measured in an in vitro translation assay. Error bars in B and C represent ±1 SD calculated from triplicate experiments.