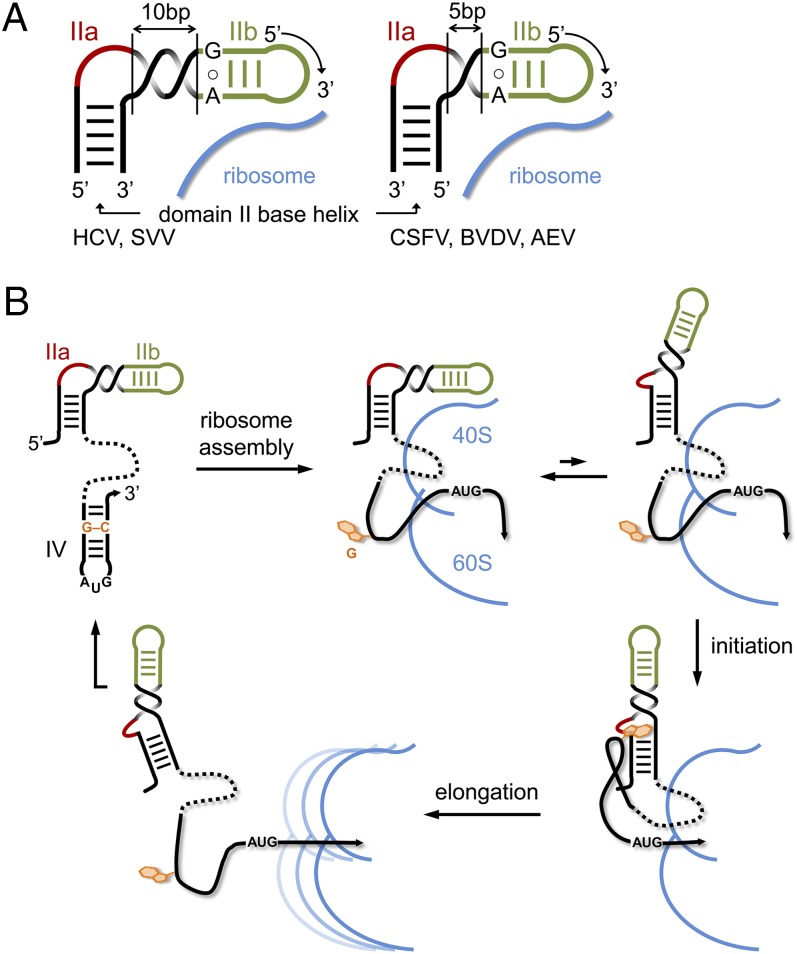
Fig. 6.
Model for the modular domain II architecture in viral IRES elements and its participation in IRES regulation. (A) For the correct positioning of the conserved hairpin loop IIb at the 40S ribosomal subunit, the bent internal loop IIa module has to be located in the 5′ or 3′ strand of the base helix, depending on the number of base pairs in the spacer segment between IIa and IIb. A spacer of 10 base pairs, as in the IRES elements of HCV and SVV, corresponds to a full turn of an A-form RNA helix, whereas a spacer of 5 base pairs provides a half turn. (B) Proposed mechanism for the regulation of IRES activity by the subdomain IIa switch through an interaction with a G residue from domain IV. Ribosome binding of the IRES entails melting of the domain IV hairpin to provide access of the viral start codon to the decoding site, which in turn unmasks the trigger G residue that facilitates IRES release during initiation by capture of the IIa extended state. After the ribosome transitions to elongation, the unbound IRES may refold to the original state.