Fig. 4.
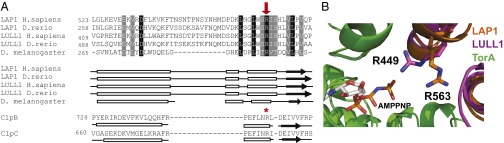
Conserved Arg residue in LAP1 and LULL1 aligns with the Arg finger of other AAA+ proteins. (A) Alignment of LAP1 and LULL1 amino acid sequences to ClpB and ClpC using predicted structural homology. LAP1 and LULL1 sequences from Homo sapiens (NP_056417 and NP_001186189), Danio rerio (NP_001017552 and XP_001339602), and Drosophila melanogaster (NP_649149) were aligned to each other using ClustalW and formatted using BoxShade; black boxes indicate strict amino acid agreement, and gray boxes indicate similar amino acid agreement. Predicted secondary structures were generated and aligned to Thermus thermophilus ClpB and Bacillus subtilis ClpC using HHPred; boxes represent helices, arrows indicate sheets, and lines indicate random coils. A conserved Arg from LAP1 (H. sapiens R563) and LULL1 (H. sapiens R449) aligns with ClpB’s and ClpC’s AAA-2 Arg finger (R747 and R704, respectively) and is indicated with a red arrow and a red star. (B) Zoomed-in view of LAP1 and LULL1’s Arg fingers in proximity to AMPPNP-bound TorA (green). LAP1 R563 (orange) and LULL1 R449 (magenta) are shown.
