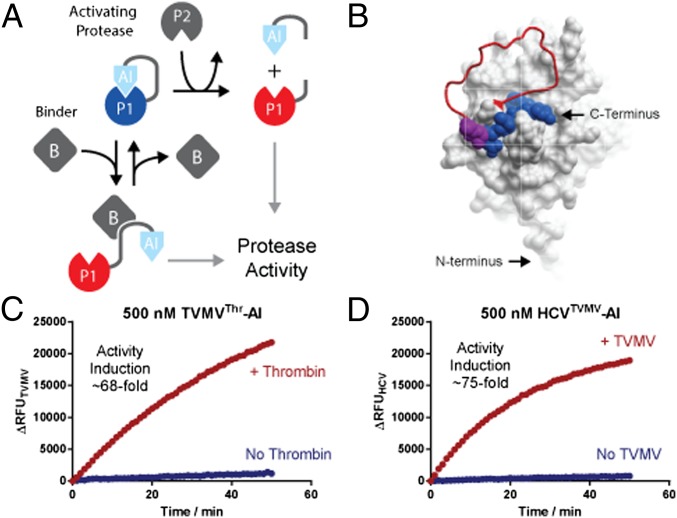
Fig. 1.
Principle design of protease-based signal transducers and ligand sensors. (A) An elementary signal transducer is created by connecting a protease domain (P1) to an autoinhibitory (AI) domain that binds and blocks the active site of the protease. Proteases are activated by dislodging the bound inhibitor from the active site through a conformational rearrangement of the linker. In the simplest case, such a rearrangement constitutes cleavage by an activating protease (P2), which separates the AI domain from the transducer protease and thus relieves autoinhibition to activate the transducer protease. Alternatively, a conformational rearrangement can be mediated through a ligand (B) binding event stabilizing the AI-domain away from the active site. (B) Surface representation of TVMV protease (Protein Data Bank ID code 3MMG) carrying the C-terminal extension (shown as red worm) containing a cleavage site for thrombin and a product peptide bound to the active site (shown in blue space filing representation). The position P5 and P6 of the substrate peptide that was varied in the library are shown as space filling magenta alanines. (C and D) Time-resolved traces of protease activities of TVMV- and HCV-based signal transducers in the presence or absence of thrombin and TVMV, respectively. Protease activities were assayed using 500 nM protease and 5 µM of quenched fluorescent substrate peptide.