Figure 4.
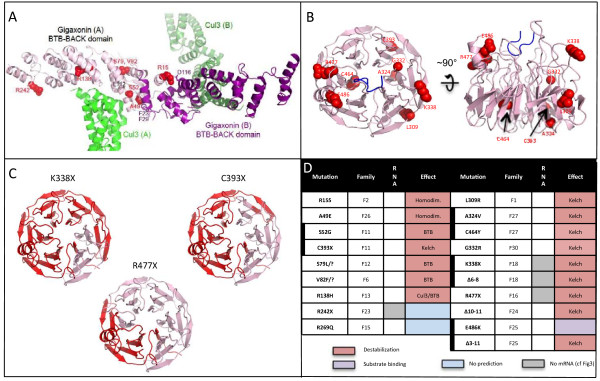
Structural modeling of Gigaxonin and predicted destabilization due to mutations. A Structural model of the homodimer BTB-BACK domain of Gigaxonin (purple A, B), in complex with with Cul3 (green A, B). Patient mutations lying in this domain are represented in red. B Representation of the top and side views of a structural model for the 6-bladed β-propeller KELCH domain of Gigaxonin. Mutations found in patients are represented in red. C Top view of the structural model for the KELCH domain of Gigaxonin, with regions deleted by the indicated truncation mutants shown in red. D Summary of the effects predicted from the modelization of Gigaxonin for all patients included in the study. Heterozygous mutations are represented by a thick vertical bar. Most of the mutations are predicted to destabilize Gigaxonin (red), whereas one of them would impair substrate binding (purple). The effect of two mutations could not be determined by the 3D model (blue).
