Figure 5.
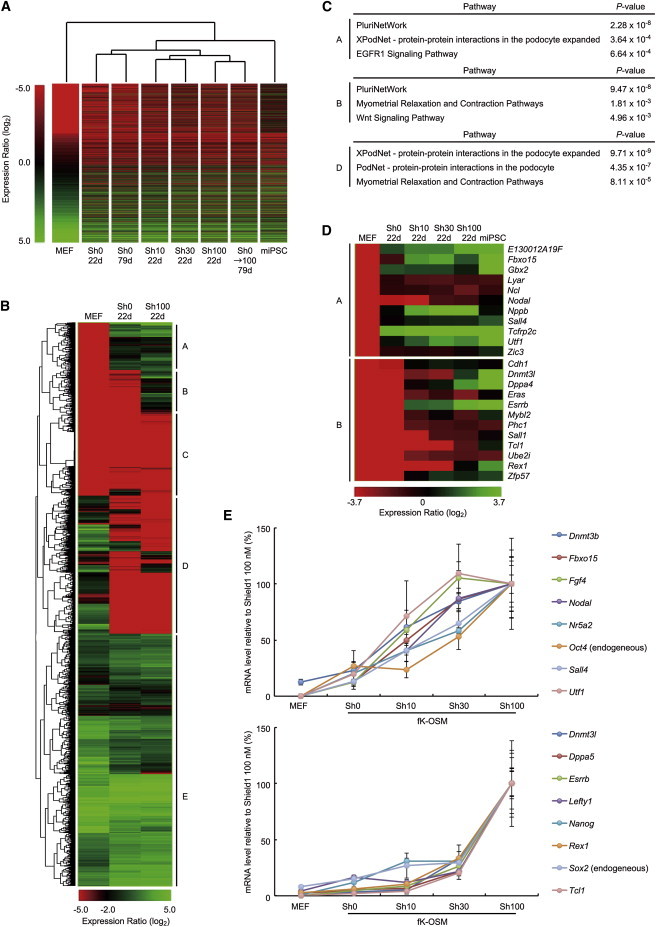
Global Gene Expression Profiles of Paused iPSCs
(A) Hierarchical clustering of paused iPSCs based on gene expression profiles of the differentially expressed genes (DEGs) between MEFs and miPSCs (p < 0.05). Analyzed samples include cells induced by SeVdp(fK-OSM) at the indicated concentrations (0, 10, 30, or 100 nM) of Shield1 for 22 or 79 days. Cells that resumed reprogramming (Sh0→100 79d) were first induced by SeVdp(fK-OSM) in the absence of Shield1 for 25 days and then cultured in the presence of 100 nM Shield1 before collection at day 79.
(B) Hierarchical clustering of DEGs. DEGs between MEFs and miPSCs (p < 0.05) were clustered based on gene expression profiles of the indicated cells and classified into five groups (group A–E).
(C) Enriched pathways in the genes of groups A, B, or D.
(D) Heatmap of the expression profiles of PluriNetWork genes clustered in group A or B.
(E) Two distinct expression profiles of pluripotency-related genes at increasing concentrations of Shield1. SeVdp(fK-OSM)-infected cells were cultured in the presence of indicated concentrations of Shield1, and the cells were collected at day 27. The mRNA levels of pluripotency-related genes were determined by quantitative RT-PCR. Data represent mean ± SEM of three independent experiments.
