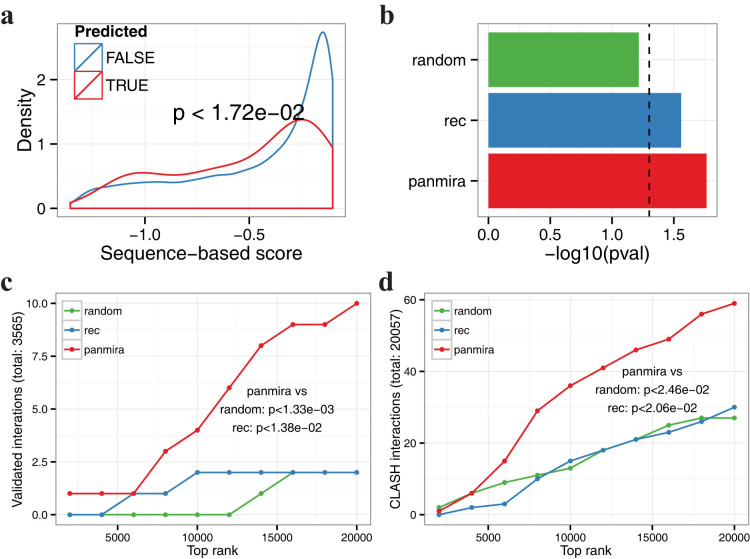
Figure 2. Validation of positive recurrent interactions.
(a) The distribution of the sequence-based scores from miRanda-mirSVR are compared between the positive recurrent interactions predicted by PanMiRa and the remaining pairs. The more negative the scores the stronger implication of a true interaction based on the sequence-based features. P-value indicated above was computed by one-sided Wilcoxon rank-sum test. (b) The same test was performed for the interactions detected by randomly shuffled posteriors and REC. The resulting significance levels in terms of -log10(p-value) are compared among the three methods. The dash line indicates the significance cutoff of p-value = 0.05. (c) & (d) Validated interactions from miRTarBase and CLASH-detected chimera are counted as a function of the top rankings from 2000–20,000 with 2000-interval. P-values indicated above were computed by comparing PanMiRa with random/REC using Wilcoxon rank-sum test.