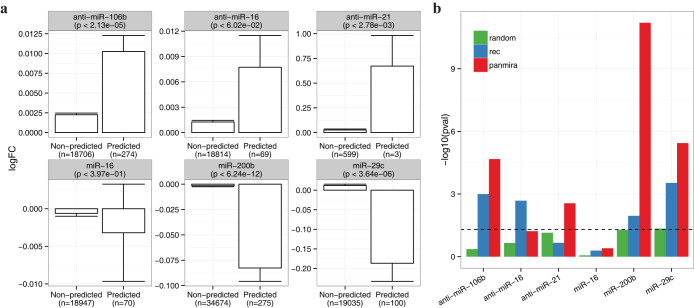
Figure 3. Functional validation using public miRNA perturbation data.
We obtained from public domains six miRNA perturbation microarray datasets measuring the expression fold-changes followed by miRNA inhibition (anti-miR-106b/16/21) or miRNA overexpression (miR-16/200b/29c). (a) Average log fold-change due to a miRNA perturbation, where the p-values in the parentheses indicate the statistical significance from one-sided Wilcoxon rank-sum test and the number of non-predicted and predicted targets are indicated at the x-axis. Error bars indicate one standard deviations from the mean. (b) The same tests were performed for the interactions detected by randomly shuffled posteriors and REC. The statistical significance levels -log10(p-value) were compared among the three methods. The dash line indicate the standard cutoff of p-value = 0.05.