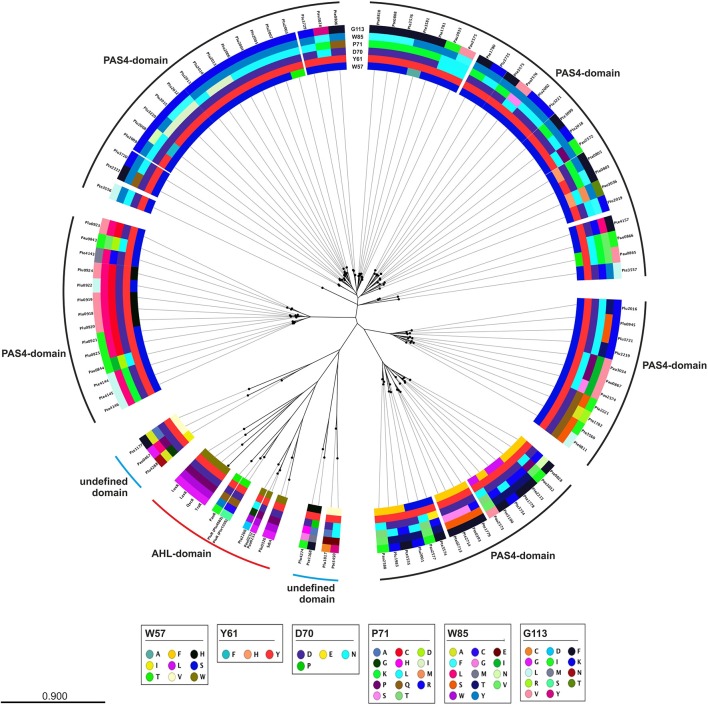
Figure 3.
Phylogenetic tree of the LuxR solos present in P. luminescens, P. temperata, and P. asymbiotica. Protein sequences of the overall LuxR solos of the three Photorhabdus species, of LuxR from Vibrio fischeri, of TraR from Agrobacterium tumefaciens, of SdiA from Escherichia coli as well as of QscR and LasR from Pseudomonas aeruginosa were aligned and a phylogenetic tree was generated. Based on this alignment the different amino acid motifs at the six conserved positions were identified by deviation from the WYDPWG-motif in the SBD of AHL-sensors. A special focus on the amino acids at positions of the WYDPWG-motif is shown from the inner to the outer circle. The amino acid W57, with respect to TraR, is marked in brown, Y61 in red, D70 in purple, P71 in dark red, W85 in pink and G113 in light pink, however substitutions within this positions are marked in different colors. LuxR-type proteins with an AHL-binding domain are marked with a red dash, LuxR-type proteins with a PAS4-domain are marked with a black dash and LuxR-type proteins with a N-terminal yet undefined domain are marked with a blue dash. Alignment and radial phylogenetic tree was generated with the CLC Mainworkbench 7 (CLC Bio Qiagen, Hilden, Germany). The scale bar indicates the length of the branches.