Abstract
Nucleic acids have been used to create diverse synthetic structural and dynamic systems. Toehold-mediated strand displacement has enabled the construction of sophisticated circuits, motors, and molecular computers. Yet it remains challenging to demonstrate complex structural reconfiguration in which a structure changes from a starting shape to another arbitrarily prescribed shape. To address this challenge, we have developed a general structural reconfiguration method that utilizes the modularly inter-connected architecture of single-stranded DNA tile and brick structures. The removal of one component strand reveals a newly exposed toehold on a neighboring strand, allowing us to remove regions of connected component strands without needing to modify the strands with predesigned external toeholds. Using this method, we have reconfigured a two-dimensional DNA rectangle canvas into diverse prescribed shapes. We also used this method to reconfigure a three-dimensional DNA cuboid.
Keywords: single-stranded tiles, DNA bricks, strand displacement
Self-assembly of nucleic acids (DNA and RNA) has produced diverse synthetic structures.[1–29] In particular, DNA origami[9, 12–15, 19, 20, 23] and single-stranded tile (SST) and bricks[21, 22, 24, 25] have enabled construction of megadalton discrete structures with arbitrarily prescribed shapes. In parallel, researchers have used strand displacement to demonstrate the construction of dynamic systems[30] such as switches,[31] walkers,[6, 32, 33] circuits,[34, 32, 35] and triggered assembly systems[36, 32] that can go through multiple states of different configurations, either in a directed or autonomous fashion.
Recently, researchers have combined DNA strand displacement techniques with DNA structural assembly methods to create reconfigurable and/or reversible structures. For example, DNA origami boxes[13] and clamshells[20] are reconfigured from a closed to an open state via toehold mediated strand displacement of a few component strands in the structure. More complex reconfiguration methods have placed single-stranded toeholds at selected sites on the structure for shape transformation,[37–40] including formation of catenane derived from DNA origami Möbius strip[39] and changing fractal patterns in origami structures[40].
However, it still remains challenging to develop a general framework for complex structural reconfiguration, in which a structure changes from a particular starting shape to another arbitrarily prescribed shape. To address this challenge, we have developed a method based on the modularly inter-connected architecture of single-stranded DNA tile and brick structures. The removal of one component strand reveals a newly exposed toehold on a neighboring strand, allowing us to remove connected regions of component strands without the need to modify them with predesigned external toeholds. It is worth noting the method applies only to SST/brick-based structures but not origami-based structures because successful reconfiguration relies on the modular architecture.
Using this method, we demonstrate that a two-dimensional rectangular DNA SST canvas[21, 25] can be reconfigured to many different shapes including two full sets of alphabet (one carved in intaglio, with the cavity forming the letters, and the other in relief, with DNA forming the letters). We also show that individual carved pieces can be re-assembled to form the original canvas and be subjected to a second round of reconfigurations. Lastly, this molecular carving concept has also been applied to a three-dimensional DNA brick cuboid structure[22]. Overall, we demonstrate that SST and brick reconfiguration is a robust and modular method for engineering complex structural reconfiguration, with resolution at the scale of the component strand (e.g. 3 nm × 7 nm for a typical 42 nt SST[21]).
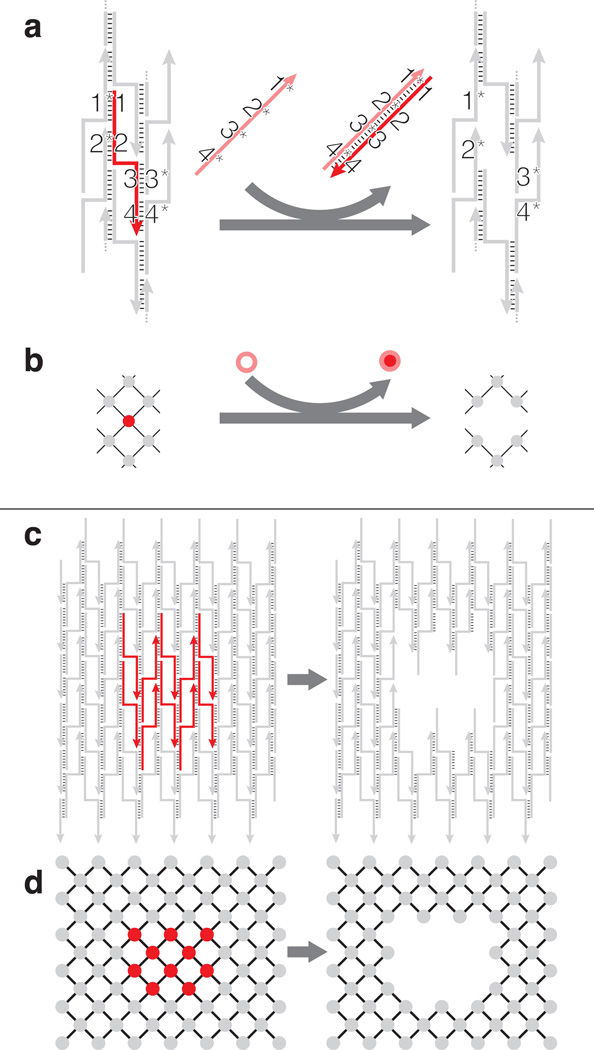
In an SST structure[21, 25], each strand typically has four binding domains that are complementary to four different neighboring strands, as depicted in Figure 1a, c. A self-assembled rectangular SST structure can be viewed as a “molecular canvas”, and depicted as an interaction graph (Figure 1b, d): each node represents a component strand and each edge represents the binding interaction between two strands or nodes. Removal (i.e. “carving”) of a component strand (red) is achieved by introducing a “carving strand” (salmon) that is fully complementary to the component strand (Figure 1a, b). Note that unlike previous strand-displacement work, our scheme does not use an external toehold[31] (The detailed molecular mechanism will be discussed later). Displacement of multiple component strands with corresponding carving strands can be used to reconfigure the canvas to a prescribed shape (Figure 1c, d). Moreover, because each component strand can be modularly removed, it is possible to create a combinatorially large number of distinct shapes.
Figure 1.
Schematics of structural reconfiguration from an SST canvas. a) A strand diagram of strand displacement based SST structural reconfiguration and b) the associated interaction graph. The strand/node to be displaced is highlighted in red with four domains 1, 2, 3 and 4 complementary to domains 1*, 2*, 3* and 4* of the neighboring strands, respectively. When introduced to the system, a full complementary strand 4*-3*-2*-1*, in salmon, forms a duplex with the red target component strand to displace the target off the canvas. c) A strand diagram of strand displacement for an 8 helix (H) × 10 turns (T) canvas and d) the associated interaction graph. Strands or nodes high- lighted in red, as shown on the left, depict the subset of component strands to be displaced. The carved structure is shown on the right.
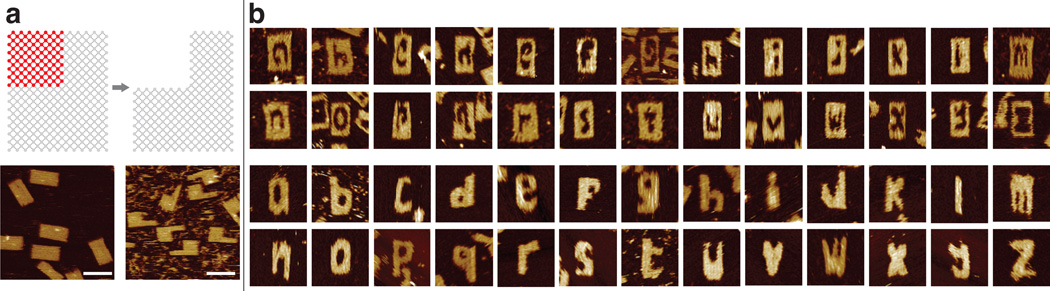
The canvas[25] was made by self-assembly of 375 distinct component strands in 15 mM Mg2+ buffer at 48°C overnight, with 30% yield as analyzed by 2% native gel electrophoresis (Figure S4). The structures showed expected morphology and dimensions under atomic force microscopy (AFM) imaging (Figure 2a, bottom left).
Figure 2.
Alphabet sets reconfigured from a rectangular SST canvas. a) Interaction graphs (top) and AFM images (bottom) of the 24H × 29T canvas used in this study (left) and its reconfiguration into a rectangle with a missing corner (right). Scale bars: 100 nm. b) AFM images of alphabet carved in intaglio (top) and relief (bottom). Each image is 150 nm × 150 nm in size. See Figure S4 and Figure S5 for agarose gel electrophoresis results.
To reduce overall time and potential human errors in picking and mixing carving strands, we wrote a computer program to design target shapes. The program takes the desired shape as input and then directs a robotic liquid handler to select the appropriate subset of strands from a master library of 375 carving strands. These carving strands were then applied to the canvas solution in an equimolar ratio for an overnight incubation at 35 or 45°C to produce the carved shapes. Figure 2a shows an example of carving a corner off the canvas (the detailed carving mechanism will be discussed later in this article). Figure 2b shows the AFM images of the carved shapes of two full sets of alphabet in intaglio and relief. The reconfigured structure in Figure 2a has a gel yield of 50% (Figure S18) and an AFM yield of 99% (N = 202; see Figure S30). Note that such a high AFM yield is not typical for the more complex alphabet structures that we carved (Figure S4 and S5, gels; Figure S6 to S15, AFM images).
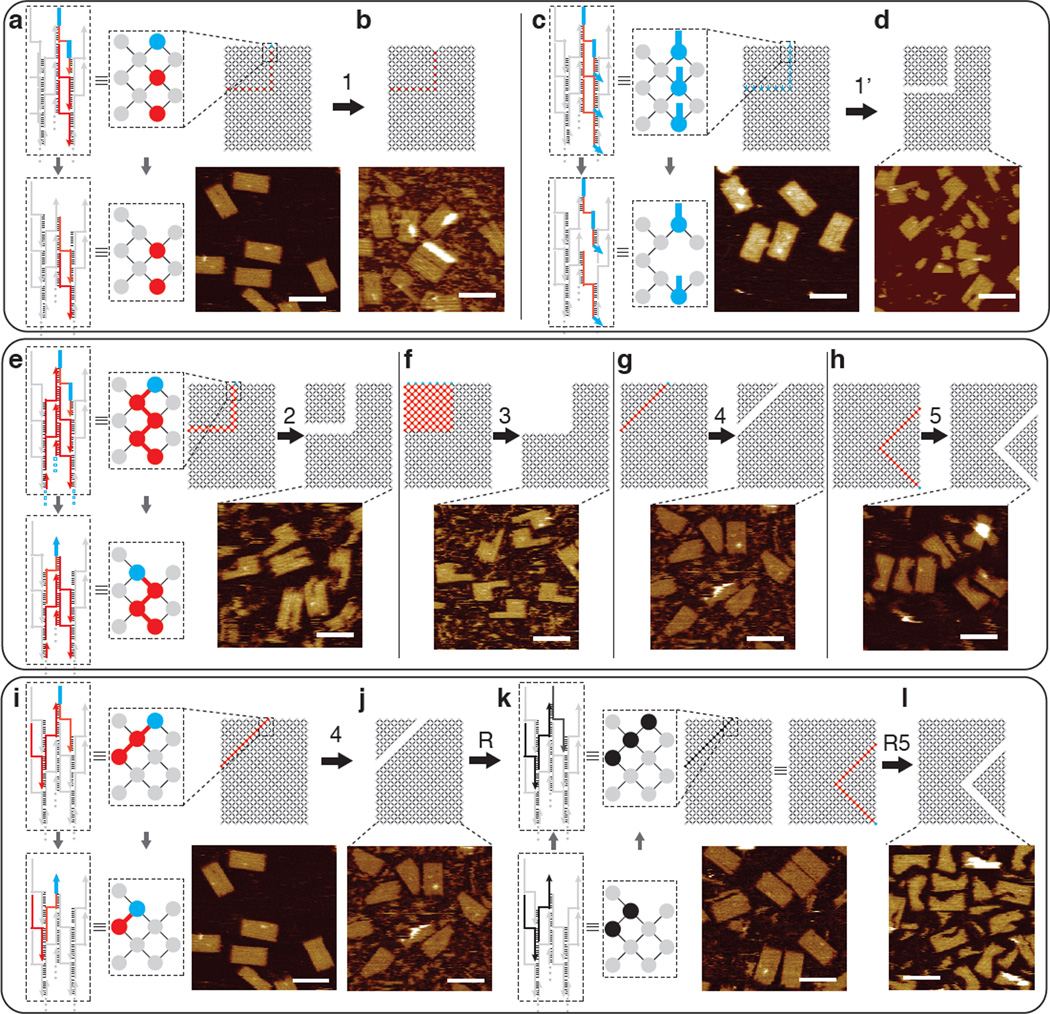
Unlike most previous strand displacement based dynamic systems, our carving scheme does not use predesigned external toeholds to initiate strand displacement. We thus conducted a set of experiments to study the effects of external toeholds on carving (Figure 3, patterns 1 and 1’ highlighted in red). We designed a different canvas where the component strands in the carving pattern have (blue) external toeholds (Figure 3c, d, pattern 1’). After overnight carving at 45°C, gel electrophoresis showed minimal carving for the toehold-free canvas (Figure 3a,b, pattern 1; yields around 0%, see Figure S16) but significant carving for the canvas with external toeholds (Figure 3c,d; yields ranging from 19–36%, see Figure S16). AFM images were consistent with the gel results: the carving on a canvas without pre- designed external toeholds rarely reached completion (~14% yield, Figure S27), while the carving on a canvas with external toeholds leads to higher percentage of completion (~81% yield, Figure S28). Note that AFM yields appeared higher than gel yields, which might reflect that partially carved structures co-migrated with intact canvas on the gel, but broke apart under AFM (e.g. during deposition or imaging process).
Figure 3.
Diagrams and AFM images of mechanism study. For all panels, top: interaction graphs with the carving pattern highlighted in red and blue; bottom: AFM images (scale bars: 100 nm). Top panel depicts carving from pattern 1 without predesigned external toeholds (a, b) and pattern 1’ with predesigned external toeholds (c, d). Middle panel (e–h) depicts carving patterns 2–5. Bottom panel (i–l) depicts reversibility of carving: reconfiguration from canvas to pattern 4, to re-assembly, and to pattern R5 (identical to pattern 5). The strand diagram and interaction graph boxed by the dashed lines show the reconfiguration mechanism at the zoomed-in fraction of the canvas (in a), c), e), i) and k)). Grey depicts common components; red depicts the strands without external toeholds to be carved; blue marks the presence of an exposed single-stranded toehold; black indicates the introduction of strands for re-assembling the carved canvas pieces. See SI Sect. S3.1 and S3.3 for detailed study of carving yields.
We next designed a second set of experiments to test five distinct connection patterns of component strands to be carved (carving patterns) in a canvas without external toeholds. The carving samples ([carving strands]: [component strands]: 1:1, 2:1, or 3:1) from patterns 1–5 were incubated at 45°C overnight before agarose gel electrophoresis and AFM imaging. When the sample from carving pattern 1 is compared with the ones from patterns 2–5, significantly lower carving yields were observed in gel electrophoresis and AFM (see SI Sect. S3.1 and Sect. S3.3 for detailed yield study).
A component strand to be carved is denoted by a red or blue node, with the blue node indicating the presence of unpaired single-stranded domain(s). Each node in carving pattern 1 (Figure 3a, b) is disjoint from any other node in the pattern. In contrast, in each of patterns 2–5 (Figure 3e–h), the nodes to be carved are fully connected such that a path consisting only of nodes to be carved exists between any two nodes within the carving pattern.
Additionally, we tested the reversibility of this structural re- configuration by adding back the displaced components following the carving of pattern 4 (Figure 3j, k, pattern R). This 45°C overnight reaction was sufficient to glue back the two carved out pieces almost seamlessly with a yield around 89% by gel and 78% by AFM (Figure 3k, S18 and S33). The re-assembled canvases were then subjected to a second round of carving (pattern R5; Figure 3l), which resulted in yields similar to those obtained directly from carving the original canvas (gel yield of 58% and AFM yield of 72%, Figure S18, S34).
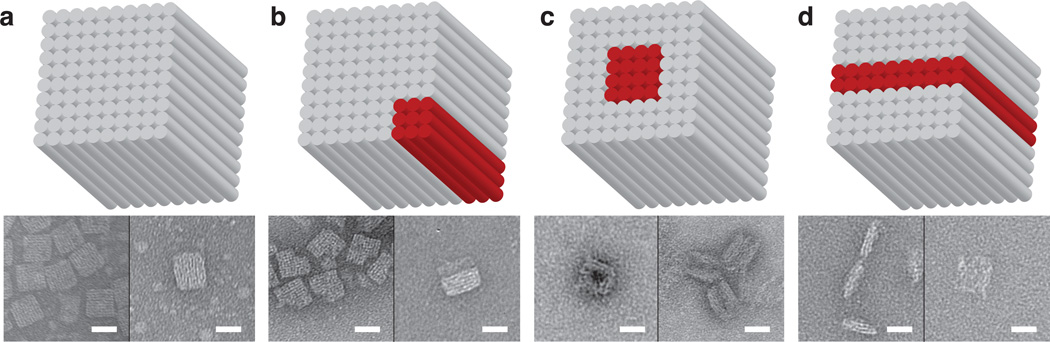
We then applied this carving method to 3D DNA brick cuboids. Using a 10 helix × 10 helix × 80 base-pair structure (Figure 4a, top) that we reported previously[22], we tested multiple carving patterns. We used a reaction temperature (28°C) lower than the 2D carving, since 3D structures contain 8 nt binding domains and are less thermally stable than the 2D structures which contain 10 or 11 nt binding domains. The successful results of carving a corner off (Figure 4b, top) or a tunnel through (Figure 4c, top) a cuboid, and carving the cuboid into two halves (Figure 4d, top) are shown in agarose gel electrophoresis (Figure S37) and Transmission Electron Microscopy (TEM) (Figure 4, bottom). Because of the limited thermal stability and the limited accessibility of majority of the component strands, the structural reconfigurations of 3D DNA structures are much more difficult to realize in comparison to the 2D counterpart and obtaining fine features is even more challenging. Since finding a temperature high enough for fast strand displacement but low enough (<30°C) to maintain structural stability is challenging, we were only able to realize a few cases of successful carving with coarse resolution.
Figure 4.
Structural reconfiguration from a 3D cuboid. Top panel, cylinder model of the cuboid (red cylinders denote the ones to be displaced); bottom panel, TEM images (scale bar: 20 nm). a) Cuboid before structural reconfiguration. b) Carving a corner from the cuboid. c) Carving a tunnel through the cuboid. d) Carving the cuboid into two halves.
Carving may not appear to be thermodynamically favorable, since a component strand in the canvas structure that is fully paired with its neighbors has the same designed number of hybridized bases as when it is bound to its fully complementary carving strand (after carving). However, carving a component strand off may help to alleviate the electrostatic repulsion that results from the closely packed neighboring DNA duplexes in the canvas structure[9, 21] and release the mechanical stress that might be accrued at the crossover points[42], thereby favoring the reaction.
Similar to typical toehold-mediated strand displacement circuits, our carving system shows leakage (carving can be initiated in a connected region even when no external toehold is present), which we utilize to initiate carving. However, it is conceivable that under other environment conditions (e.g. at room temperature over a shorter period of time rather than at the elevated temperature [35– 45°C] overnight used in our experiments), such leakage could be mitigated and carving may only start at a site with an external toehold. Such a system would be analogous to a strand displacement circuit embedded in a nanostructure (the canvas), where the strand displacement cascade is directly coupled with structural change at a single strand resolution. This system would provide a platform to explore the rich interplay between structure, dynamics, and computation. For example, it would be interesting to design spatial logic gates and circuits: a multi-input AND gate could be designed as a carving path with the component strands along the path serving as the inputs and the strand at the end of the path serving as the output; a 2-input OR gate could be designed as two converging paths with one strand on each path serving as the inputs and the strand at the merging point serving as the output. In the latter case, when multiple strands along each path are used as inputs, these two converging paths together implement a disjunctive normal form formula ([A1 AND A2 AND … AND Am] OR [B1 AND B2 AND … AND Bn]).
Supplementary Material
Footnotes
We thank David Zhang and Yonggang Ke for discussion; Michelle K. Vhudzijena for technical assistance. This work is supported by an Office of Naval Research (ONR) Young Investigator Program Award (N000141110914), ONR grants (N000141010827 and N000141310593), a National Science Foundation (NSF) Faculty Early Career Development Award (CCF1054898), an NSF Expedition in Computing Award (CCF1317291), NSF grants (CCF1162459, CMMI1333215), a National Institutes of Health (NIH) Director’s New Innovator Award (1DP2OD007292) and a Wyss Institute for Biologically Engineering Faculty Startup Fund to P.Y.. L.L.O. acknowledges support from NSF through a Graduate Research Fellowship.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201xxxxxx.
Contributor Information
Bryan Wei, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115; Department of Systems Biology, Harvard Medical School, Boston, MA 02115.
Luvena L. Ong, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115 Harvard-Massachusetts Institute of Technology (MIT) Division of Health Sciences and Technology, MIT, Cambridge, MA 02139.
Jeffrey Chen, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115.
Alexander S. Jaffe, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115
Peng Yin, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115; Department of Systems Biology, Harvard Medical School, Boston, MA 02115.
References
- 1.Seeman NC. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 3.Fu TJ, Seeman NC. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 4.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 5.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 6.Sherman WB, Seeman NC. Nano Lett. 2004;4:1203–1207. [Google Scholar]
- 7.Chworos A, Severcan I, Koyfman AY, Weinkam P, Emin Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 8.Park SH, Pistol C, Ahn SJ, Reif JH, Lebeck AR, Dwyer C, Labean TH. Angew. Chem. Int. Ed. 2006;45:735–739. doi: 10.1002/anie.200503797. Angew. Chem. 2006, 118, 749–753. [DOI] [PubMed] [Google Scholar]
- 9.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 10.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 11.Yin P, Hariadi R, Sahu S, Choi HMT, Park SH, LaBean TH, Reif JH. Science. 2008;321:824–826. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 12.Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H. Nano. Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 13.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 14.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietz H, Douglas SM, Shih WM. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng JP, Birktoft J, Chen Y, Wang T, Sha RJ, Constantinou P, Ginell S, Mao C, Seeman NC. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. Nat. Chem. 2010;2:772–779. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 19.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 20.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 21.Wei B, Dai M, Yin P. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ke Y, Ong LL, Shih WM, Yin P. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han D, Pal S, Yang Y, Jiang S, Nangreave J, Liu Y, Yan H. Science. 2013;339:1412–1415. doi: 10.1126/science.1232252. [DOI] [PubMed] [Google Scholar]
- 24.Myhrvold C, Dai M, Silver PA, Yin P. Nano Lett. 2013;13:4242–4248. doi: 10.1021/nl4019512. [DOI] [PubMed] [Google Scholar]
- 25.Wei B, Dai M, Myhrvold C, Ke Y, Jungmann R, Yin P. J. Am. Chem. Soc. 2013;135:18080–18088. doi: 10.1021/ja4062294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C, Liu Y, Rinker S, Yan H. Chem Phys Chem. 2006;7:1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 27.Shih WM, Lin C. Curr. Opin. Struct. Biol. 2010;20:276–282. doi: 10.1016/j.sbi.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leontis NB, Lescoute A, Westhof E. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seeman NC. Annu. Rev. Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang DY, Seelig G. Nature Chemistry. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 31.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 32.Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 33.Omabegho T, Sha R, Seeman NC. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seelig G, Soloveichik D, Zhang DY, Winfree E. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 35.Qian L, Winfree E. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 36.Dirks RM, Pierce NA. Proc. Natl. Acad. Sci. USA. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan H, Zhang Z, Shen X, Seeman NC. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Ke Y, Lin C, Yan H, Liu Y. Chem. Comm. 2008:4318–4320. doi: 10.1039/b807933a. [DOI] [PubMed] [Google Scholar]
- 39.Han D, Pal S, Liu Y, Yan H. Nat. Nanotechnol. 2010;5:712–717. doi: 10.1038/nnano.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, Nangreave J, Liu Y, Yan H. Nano Lett. 2012;12:3290–3295. doi: 10.1021/nl301399z. [DOI] [PubMed] [Google Scholar]
- 41.Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 42.Maiti PK, Pascal TA, Vaidehi N, Goddard WA. Nucleic Acids Res. 2004;32:6047–6056. doi: 10.1093/nar/gkh931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.