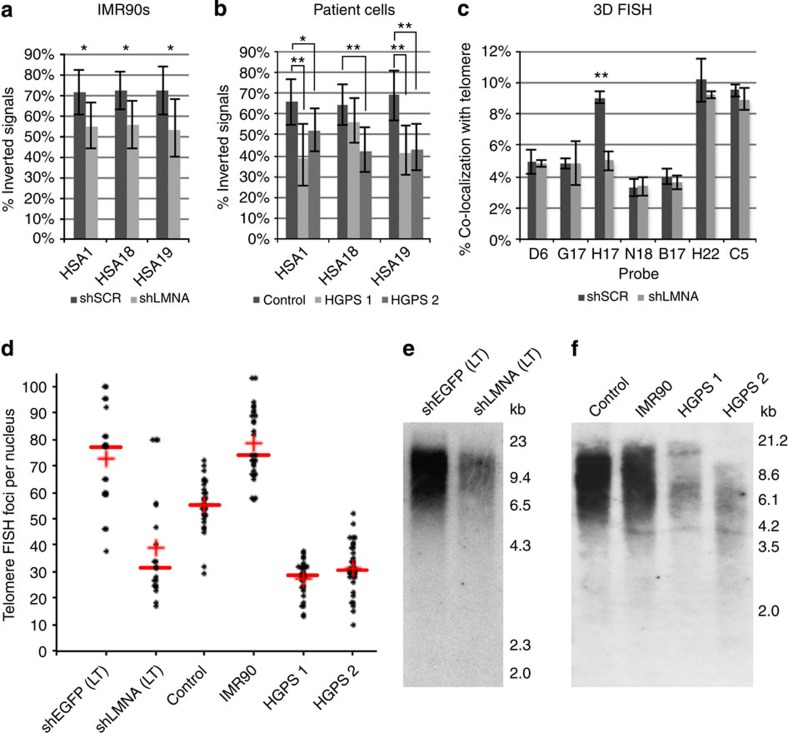
Figure 4. Lamin A/C stabilizes chromosome-end structure and is necessary for telomere stability.
The frequency of inverted chromosome-end structure was quantified for HSA1, HSA18 and HSA19. For each chromosome, the positioning of a telomeric probe signal was compared with the positioning of the indicated subtelomeric probe signal. This analysis was performed for (a) IMR90s treated with shLMNA or shSCR (n=45–89 signals per probe set) *P<0.05, Student s t-test and (b) fibroblasts from a healthy patient (control) and two different HGPS patients (HGPS 1 and HGPS 2; n=39–83 signals per probe set). *P<0.05, **P<0.01, Student’s t-test. (c) Quantification of IMR90 interphase nuclei treated with shLMNA or shSCR for co-localization between BACs on HSA1 (see Fig. 2a) and telomere signal. Results are mean±s.e.m. (n=3). **P<0.01, Student’s t-test. (d) Quantification of the number of telomeric FISH foci per nucleus in shLMNA(LT) or shEGFP(LT) IMR90s, as well as control or HGPS patient cells. The red line and cross indicate the data set median and mean, respectively. (e) Southern blot analysis of telomeric DNA size and abundance in shLMNA(LT) or shEGFP(LT) IMR90s. (f) Southern blot analysis of telomeres from control patient cells and HGPS patient cells, telomeric DNA from IMR90s is included for comparison.