Guiu et al. use ChIP-on-chip analysis for the Notch partner RBPj, using embryonic tissue from the aorta-gonad-mesonephros region to identify potential novel Notch target genes involved in HSC emergence. They show that c-MYC–responsive gene Cdca7 is expressed in different HSC and progenitor subpopulations and that CDCA7 is important for maintaining the undifferentiated phenotype. Cdca7 acts downstream of Notch in HSCs in zebrafish, mouse, and human, indicating a highly conserved Notch/RBPj/Cdca7 axis in hematopoietic development.
Abstract
Hematopoietic stem cell (HSC) specification occurs in the embryonic aorta and requires Notch activation; however, most of the Notch-regulated elements controlling de novo HSC generation are still unknown. Here, we identify putative direct Notch targets in the aorta-gonad-mesonephros (AGM) embryonic tissue by chromatin precipitation using antibodies against the Notch partner RBPj. By ChIP-on-chip analysis of the precipitated DNA, we identified 701 promoter regions that were candidates to be regulated by Notch in the AGM. One of the most enriched regions corresponded to the Cdca7 gene, which was subsequently confirmed to recruit the RBPj factor but also Notch1 in AGM cells. We found that during embryonic hematopoietic development, expression of Cdca7 is restricted to the hematopoietic clusters of the aorta, and it is strongly up-regulated in the hemogenic population during human embryonic stem cell hematopoietic differentiation in a Notch-dependent manner. Down-regulation of Cdca7 mRNA in cultured AGM cells significantly induces hematopoietic differentiation and loss of the progenitor population. Finally, using loss-of-function experiments in zebrafish, we demonstrate that CDCA7 contributes to HSC emergence in vivo during embryonic development. Thus, our study identifies Cdca7 as an evolutionary conserved Notch target involved in HSC emergence.
Hematopoietic stem cells (HSCs) emerge from the major arterial vessels during embryonic development. Embryonic vascular development is closely associated with HSC generation because arteries provide the niche HSC generation and both lineages share a common endothelial progenitor (Zovein et al., 2008; Chen et al., 2009). The process by which an HSC precursor with endothelial characteristic acquires the hematopoietic identity is known as endothelial to hematopoietic transition. HSCs develop within specific cell clusters budding from the endothelium to the lumen of the dorsal aorta in the region comprised between the junctions of the vitellin and umbilical arteries (Yokomizo and Dzierzak, 2010). These hematopoietic clusters contain a variety of cells that express different cell surface markers such as c-kit or CD41 or CD45 and include those that will acquire the stemness capacity. After release into circulation, these cells are amplified in the fetal liver, giving rise to the adult HSCs.
The process of HSC generation requires the orchestration of crucial developmental pathways, including Notch and Wnt (Robert-Moreno et al., 2005; Ruiz-Herguido et al., 2012). Notch signaling regulates cell fate decisions with a central role in vascular and hematopoietic development (Bigas and Espinosa, 2012). Notch activity is first required to generate arteries, and Notch inhibition favors vein formation from the prepatterned endothelial network (You et al., 2005). Activation of Notch can be achieved by its interaction with either Delta or Jagged ligands, thus triggering the proteolytic cleavage and release of the active Notch intracellular fragment (ICN) that will induce a transcriptional response together with its nuclear partners RBPj and Mastermind (Mam). However, Notch activation during arterial determination specifically depends on the Delta4 ligand (Duarte et al., 2004; Krebs et al., 2004), whereas HSC generation in the hematopoietic clusters of the aorta-gonad-mesonephros (AGM) is mostly dependent on Jagged1 (Robert-Moreno et al., 2008). Thus, Jagged1-deficient embryos provide a unique system to study the role of Notch in embryonic hematopoiesis in a normal arterial scenario. This specific Notch function is not restricted to mammals, as it also regulates zebrafish (Burns et al., 2005) as well as Drosophila melanogaster hematopoietic development (Mandal et al., 2004; Terriente-Felix et al., 2013). In the mouse, only two direct Notch targets involved in HSC generation have been identified, but it is expected that other genes that participate in this process will also depend on Notch, as it has been shown in Drosophila (Terriente-Felix et al., 2013). In particular, Notch1 receptor signaling induces the activation of an incoherent feed-forward loop involving the Hairy and enhancer of split 1 (Hes1) repressor and the Gata2 transcription factor, which results in fine tuning of Gata2 levels and is essential to generate functional HSCs (Guiu et al., 2013). Similar regulatory loops for other Notch-dependent genes have been identified in Drosophila (Krejcí and Bray, 2007), which indicates the conservation of a mechanism that modulates context-specific targets through general Notch effectors such as Hes repressors. Genes regulated by these feed-forward regulatory loops are hard to identify in most of the screenings because once Notch is artificially stimulated or repressed, both the activating and the repressing complexes are simultaneously modified.
To identify novel HSC regulators that are targets of Notch in the AGM, we have based our strategy on (a) the identification of gene promoters that bind RBPj, (b) the selection of candidate genes by the presence of RBP binding consensus, and (c) the analysis of the expression patterns in the AGM of WT and Jag1 mutant embryos. Following this strategy, we identified Cell division cycle associated 7 (Cdca7) as a Notch/RBPj target gene that is activated in the HSC precursors in the mouse embryo and maintained thereafter in distinct undifferentiated hematopoietic populations. Notch-dependent induction of Cdca7 expression is recapitulated during early hematopoietic differentiation of human embryonic stem cells (ESCs [hESCs]), whereas down-regulation of Cdca7 in the AGM cells induces a rapid differentiation of hematopoietic progenitors. In the zebrafish embryo, knocking down Cdca7 significantly reduced HSC generation in vivo.
RESULTS
Screening for novel Notch/RBPj transcriptional targets in the AGM region
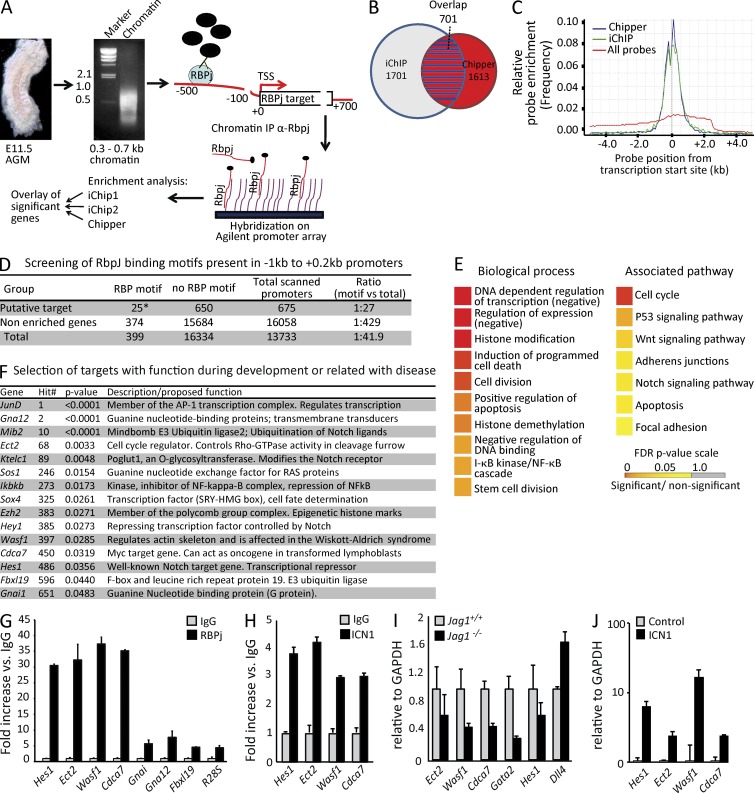
We performed chromatin immunoprecipitation (ChIP)–on-chip analysis with anti-RBPj antibodies using cross-linked chromatin from embryonic day (E) 11.5 dissected AGMs (scheme in Fig. 1 A). A set of putative targets was obtained with a stringent analysis by combining three bioinformatic tools (iChip1, iChip2, and Chipper; Fig. 1 B). Compared with the whole set of probes represented in the array (includes from −5 to 2.5 kb from the transcriptional start sites [TSSs]), RBPj-bound regions showed enrichment in the proximal promoters, including the TSSs (Fig. 1 C). When overlapping the putative RBPj/Notch targets from the three independent analyses, 701 promoter regions showed enriched RBPj recruitment relative to the input, of which 676 (675 genes and 1 miRNA) were annotated with an Ensembl gene ID (Fig. 1 B and Table S1). Our list of RBP-binding genes included two Hey/Hes family members, as well as other previously identified Notch targets such as Bcl-2, NRARP, PTEN, and Deltex2 (Table S1). Precipitation with an irrelevant Ig confirmed the specificity of the experiment (not depicted). In silico analysis of the promoter regions of identified genes showed a strong enrichment of RBPj motifs in the −1-kb region from their TSS sequence (Fig. 1 D). To investigate the possibility that Notch/RBPj-regulated genes in the AGM were clustered into specific functions or signaling pathways, Gene Ontology (GO) analysis (GO gene function and GO KEGG pathways) was performed. We found that genes identified as RBPj targets associated with DNA/chromatin regulation, cell division, and stem cell division. Remarkably, genes associated to the Wnt and Notch pathways were also significantly enriched in the group of RBPj targets (Fig. 1 E). By literature mining, we found that several of the identified genes played a recognized function in embryonic development, cancer, and/or hematopoiesis (Fig. 1 F). These genes include the transcription factors SRY-related HMG-box-4 (Sox4) and homeobox A1 (HoxA1), the cell cycle regulator Cdca7, the Notch receptor modulator Ktelc1 (Rumi), the Delta ubiquitin ligase Mindbomb2 (Mib2), and the canonical Notch targets Hes1 and Hey1.
Figure 1.
ChIP-on-chip analysis of E11.5 AGMs. (A) Schematic representation of the ChIP-on-chip procedure. (B) 701 overlapping promoter regions (red with blue stripes) between iChip (blue circle) and Chipper (red circle) software were identified. (C) Comparing the relative probe enrichment of identified RBPj targets by Chipper and iChip to all probes. (D) RBPj-binding motif scanning (TGGGAAANT) in a range of −1 to 2 kb from the TSS showed significantly more binding sites in the 675 promoter regions with an Ensembl ID than all of the promoter regions (*, P = 0.021; χ2 = 5.259). (E) GO biological process (GO-BP) and pathway (KEGG) analysis in the 676 overlapping group. (F) Selection of putative RBPj targets showing the gene symbol, hit number, and enrichment p-value. (G) qPCR analysis for RBPj recruitment to the selected genes in an independent ChIP from E11.5 AGMs (n = 3). (H) Recruitment of ICN1 to the selected genes, assessed by ChIP from E11.5 AGMs (n = 3). (I) qRT-PCR analysis on cDNA of E10.5 dissected AGMs from Jag1+/+ and Jag1−/− embryos from confirmed ICN1 and RBPj target genes (Ect2, Wasf1, and Hes1); Gata2 and Dll4 are shown as controls (n = 4). (J) A pool of six E10.5 AGMs from Jag1−/− was disrupted with collagenase and cultured. In this culture ICN1-IRES-GFP was transduced, and after 1 wk, infected cells (GFP+) were sorted and the indicated genes were analyzed by qRT-PCR (n = 3). Error bars represent the SD.
We further validated our ChIP-on-chip results by quantitative PCR (qPCR) analysis on an independent RBPj precipitation using primers around the enriched peaks of six randomly selected target genes. Analysis of the WAS protein family member 1 (Wasf1), Epithelial cell transforming sequences 2 oncogene (Ect2), Cdca7, Hes1, using the 28S ribosomal RNA (R28S) as a negative control, confirmed the enrichment detected by ChIP-on-chip (Fig. 1 G). In contrast, the genes Fbx19, Gna12, and Gnai, which were also identified in the initial screening, showed very poor enrichment ratios and were therefore considered as false positives (Fig. 1 G). Because RBPj recruitment is not exclusive of genes that contain active Notch, we analyzed whether the truncated intracellular Notch1 (ICN1) was also recruited to these promoters in the AGM of E11.5 embryos. ChIP assay using specific antibodies showed that four validated RBPj target promoters (Wasf1, Ect2, Cdca7, and hes1) also recruited active Notch1 (Fig. 1 H), supporting the notion that Notch1 regulates their expression at the time of HSC emergence.
Novel identified Notch/RBPj targets are Jagged1/Notch1-dependent genes
Because HSC generation is highly dependent on the Notch ligand Jag1 (Robert-Moreno et al., 2008), we focused on the identification of Notch/RBPj targets that are misregulated in a Jag1-deficient background. We obtained RNA from E10.5 AGMs of Jag1+/+ and Jag1−/− embryos and performed expression analysis of our candidate genes. All selected Notch targets were down-regulated in the absence of Jag1, in contrast to the Notch ligand Dll4 which was up-regulated in this specific mutant (Fig. 1 I). The Notch target Gata2 was also down-regulated in the Jag1-deficient AGM as shown previously (Robert-Moreno et al., 2008). Conversely, transduction of AGM cells from Jag1−/− embryos with active ICN1 resulted in the up-regulation of all selected targets (from 2- to 16-fold) compared with the control (Fig. 1 J).
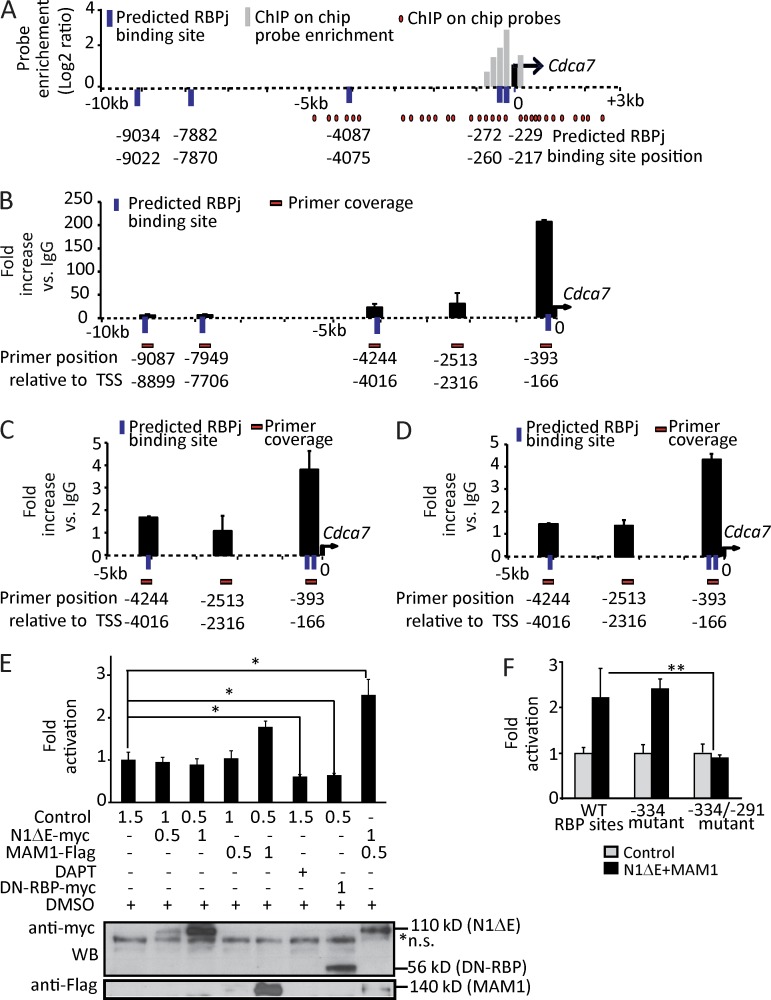
Analysis of the sequence comprising 10 kb of the 5′ regulatory regions using the Genomatix software demonstrated that putative RBPj-binding sites were present in the proximal promoter of all of the identified targets. Head-to-head dimerization of the Notch factor recruited to closely localized RBPj-binding sites (15–17 bp from core to core) is important for transcriptional regulation of specific genes such as Hes1 (Fig. S1; Arnett et al., 2010). Interestingly, we found that the Cdca7 promoter contained three putative RBPj-binding sites in the region comprised between the −5 kb and the TSS, and it contained two adjacent RBPj sites separated by a distance of 43 bp that mapped close to the TSS. This regulatory region was covered by the Chip-on-chip probes and was highly enriched in the RBPj precipitation (Fig. 2 A).
Figure 2.
Cdca7 is a direct Notch target gene through two predicted RBPj-binding sites. (A) Schematic representation of the promoter region of Cdca7 showing the array probe position (red circles), DNA (black dashed line), TSS (black arrow), putative RBPj-binding site (blue bars), and probe enrichment in log2 ratio (gray bars). RBPj-binding sites were identified with the STORM and Genomatix software scanning from the −10- to 0-kb region of each target gene. (B) ChIP with anti-RBPj. (C) ChIP with anti-N1IC. (D) ChIP with anti-MAM1. In B–D, chromatin was extracted from E10.5 dissected AGMs (n = 2). (E and F) Luciferase activity from Cdca7 promoter (−511/50 bp) WT (E) and mutant RBP sites (F) in the presence of the specified transfected plasmids into HEK-293T cells (micrograms of transfected plasmid are indicated; n = 3). In the bottom panels, Western blot of the transfected proteins is shown. Error bars represent the SD. Significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.01).
Cdca7 transcriptional activation depends on Notch signaling
We further validated the RBPj recruitment to Cdca7 using primers covering the different regions of the locus (Fig. 2 B). We confirmed that the region containing the two adjacent RBPj-binding sites (−260 and −217 bp) was highly enriched in the RBPj precipitates when normalized for the control IgG precipitation (∼206-fold) or when compared with other regions including putative binding sites (−4,087, −7,882, and −9,004 bp) in the Cdca7 gene (Fig. 2 B). Importantly, Notch1 and its coactivator Mam were also specifically recruited to the same region of the Cdca7 promoter (Fig. 2, C and D). To study whether Notch regulated Cdca7 transcription, we cloned the region comprised between −511 and 50 bp of the gene on a luciferase reporter vector and tested its response to Notch activation or inactivation in HEK-293T cells. Simultaneous expression of the ICN1 construct (N1ΔE) and Mam significantly increased the activity of the Cdca7 promoter compared with the control or with ICN1 construct alone (Fig. 2 E). Conversely, inhibition of Notch activity by DAPT or by transfection of a dominant-negative form of RBPj resulted in a significant reduction in the activity of the promoter (Fig. 2 E). Furthermore, we found that mutation of both adjacent RBP-binding sites totally abolished the Notch-dependent activation of the construct (Fig. 2 F), whereas single mutations of the distal site (−260) did not significantly affect its activity. Altogether, these results indicate that a Notch complex containing RBPj, Notch1, and Mam directly regulates the transcriptional activation of Cdca7. However, in vitro activation of the proximal promoter does not require Notch homodimerization.
Cdca7 is expressed in the aortic clusters and hemogenic precursors from AGM and hESCs
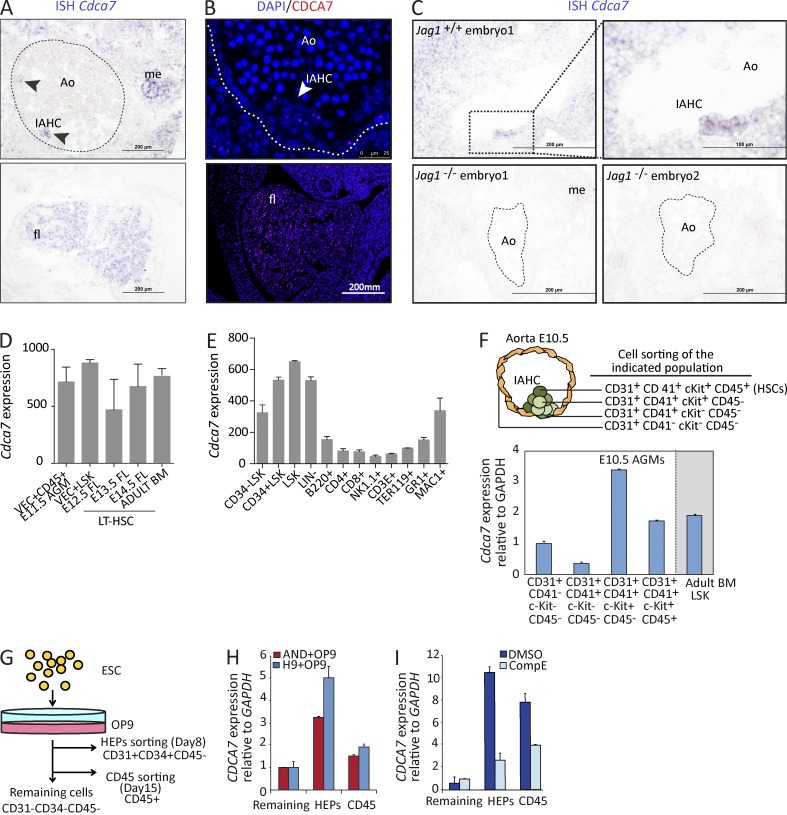
We next investigated the putative role of CDCA7 during development of the murine hematopoietic system. By in situ hybridization (ISH) of midgestation embryos, we found that Cdca7 was mainly expressed in the hematopoietic cluster–like structures in the AGM, which contain the newly formed HSCs, but also in the fetal liver (Fig. 3 A). In contrast, Cdca7 was not detected in the rest of the aortic endothelial cells, suggesting that Cdca7 is involved in hematopoietic emergence but not in arterial specification (Fig. 3 A). Similar results were obtained by immunostaining of the CDCA7 protein (Fig. 3 B). As expected, ISH analysis showed that Cdca7 expression was completely abolished in the aorta of Jag1−/− embryos (Fig. 3 C), in agreement with the qRT-PCR results (Fig. 1 I).
Figure 3.
Cdca7 is expressed in mouse and human embryonic hematopoietic progenitor cells. (A) ISH of Cdca7 on E10.5 embryos. Arrowheads indicate hematopoietic clusters in the top panel. Fetal liver (fl) staining is shown in the bottom panel. (B) E11.5 embryo immunofluorescence is shown. (C) ISH comparing E10.5 Jag1+/+ with Jag1−/− embryos. Dashed lines indicate the aortic endothelium and bars indicate the scale. Ao, aorta in the AGM region; IAHC, intraaortic hematopoietic cluster; me, mesonephros. (D–F) Cdca7 expression from the indicated purified populations from the embryo to the adult. (D and E) Gene expression from purified HSCs obtained from E11.5 AGM (VEC+CD45+), E12.5 FL (LSK), and LT-HSCs from adult BM (CD150+CD48−LSK) and different subpopulations of hematopoietic progenitors and mature cells. Data are derived from public databases. (F) Cdca7 expression on purified E10.5 AGM subpopulations and adult BM LSK. (G) Experimental design of hESC hematopoietic differentiation on OP9. (H and I) HEPs (CD31+CD34+CD45−) and CD45+ hematopoietic differentiated cells were sorted and analyzed by qRT-PCR relative to nonhematopoietic remaining cells (CD31−CD34−CD45−). Cdca7 expression was determined by qPCR (n = 3). Error bars represent the SD.
To study whether Cdca7 is differentially expressed in specific HSC populations, we took advantage of gene expression data available in public databases (GEO accession nos. GSE37000 and GSE27787). These experiments include data of purified HSCs obtained from E11.5 AGM (VEC+CD45+), E12.5 FL (LSK), and LT-HSCs from adult BM (CD150+CD48−LSK; McKinney-Freeman et al., 2012) and different subpopulations of hematopoietic progenitors and mature cells (Konuma et al., 2011). Interestingly, Cdca7 mRNA is highly expressed in HSCs from all different developmental stages (Fig. 3 D), and its expression is lower in differentiated cells from specific hematopoietic lineages (Fig. 3 E). To directly determine the expression levels of Cdca7 in different hematopoietic subpopulations of the AGM clusters at E10.5, we obtained mRNA from cells sorted using specific markers for pure endothelial (CD31+cKit−CD41−CD45−), hemogenic endothelial (CD31+cKit−CD41+CD45−), prehematopoietic (CD31+cKit+CD41+CD45−), and hematopoietic cluster cells (CD31+cKit+CD41+CD45+). Cdca7 was up-regulated in the sorted cKit+ cells, when compared with the cKit−, with ∼5 log2–fold difference (Fig. S2 A). The highest Cdca7 levels were detected in the CD31+CD41+cKit+CD45− population (Fig. 3 F), which contains the putative precursors of the HSCs. To a lesser extent, Cdca7 was also present in the population that contains the functional HSCs and hematopoietic progenitors (CD31+CD41+cKit+CD45+) and maintained in the LSK population isolated from adult BM (Fig. 3 F). Based on all of this data, we proposed that CDCA7 may play a role during endothelial to hematopoietic transition in the human embryo (Ivanovs et al., 2011). We took advantage of the hematoendothelial differentiation system using the hESC lines H9 and AND1 and two hematopoietic differentiation strategies: embryoid bodies (EBs; Fig. S2 B) and OP9 co-culture system (Fig. 3 G; Wang et al., 2004; Real et al., 2012; Bueno et al., 2013). In both conditions, we observed the appearance of hematoendothelial precursors (HEPs; CD31+CD34+CD45−), followed by the generation of hematopoietic cells (CD45+). We sorted these subpopulations, obtained mRNA, and measured the expression levels of human CDCA7 variant 2, which is homologous to the murine Cdca7 (Fig. S2 C). We found that CDCA7 mRNA was strongly enriched in the human HEPs obtained from both EB and OP9 differentiation systems, confirming the trend observed in the murine AGM cells (Fig. S2 D and Fig. 3 H). To test whether CDCA7 expression during hematopoietic differentiation of hESC was dependent on Notch activity, which is known to modulate hematopoietic differentiation of ESCs (Yu et al., 2008; Lee et al., 2013; Rafii et al., 2013), we included Notch/γ-secretase inhibitor (CompE) in our experiments. We found that compE treatment significantly reduced the levels of CDCA7 expression in both the HEP and the CD45+ populations (Fig. 3 I). Further supporting a role for CDCA7 downstream of Notch in the human hematopoietic system, we found that Notch1 is recruited to the region spanning 400 bp to the 5′ region of CDCA7 in human peripheral blood CD34+ cells (Lin, M.I., and L. Zon, personal communication).
Cdca7 maintains the undifferentiated phenotype of the hematopoietic lineage
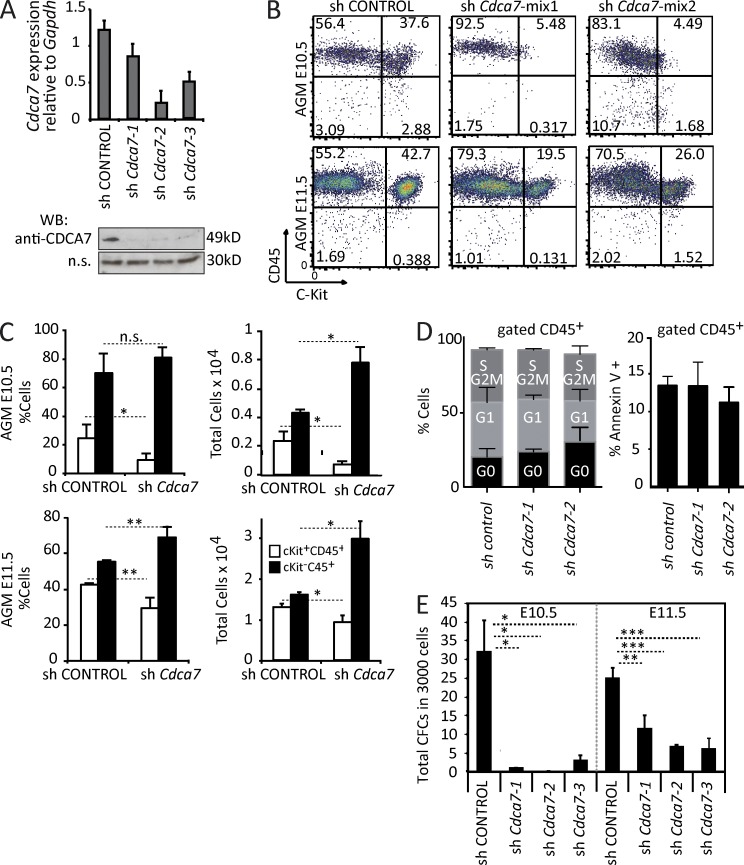
To directly test the functional requirement of CDCA7 during mouse embryonic hematopoiesis, we obtained E10.5 AGM cells and transduced them with lentiviral vectors containing shRNA targeting Cdca7 or the scrambled control (Fig. 4 A). Control cells or Cdca7 knocked down cells were cultured for 7 d in the presence of cytokines and analyzed by flow cytometry for the presence of CD45+ and cKit+ cells. In the control cultures, we obtained a mean of 30% of CD45+cKit+ double-positive hematopoietic progenitors that was reduced by eightfold after Cdca7 knockdown. In contrast, these cultures showed a forced differentiation into more mature CD45+cKit− cells. Similar results were obtained from E11.5 AGM cultures (Fig. 4 B). Reduction in the hematopoietic progenitors was also detected when total cell numbers were analyzed (Fig. 4 C). However, total cell number after culture was similar in both conditions, and we did not detect any significant effect on the cell cycle profile or the apoptotic rate in the Cdca7-depleted hematopoietic cells (Fig. 4 D). These results strongly suggest that CDCA7 regulates hematopoietic emergence and differentiation rather than survival or proliferation of existing hematopoietic cells. Furthermore, knocking down Cdca7 in AGM cells totally abolished (in E10.5 cells) or strongly reduced (in E11.5 cells) their capacity to generate lineage-specific colonic (CFCs; Fig. 4 E). These results indicate that CDCA7 has a dominant role in maintaining the undifferentiated phenotype of the newly formed HSCs and progenitors.
Figure 4.
CDCA7 is required to maintain embryonic hematopoietic progenitors. (A) shCdca7-1, shCdca7-2, and shCdca7-3 were tested in 32D myeloid progenitor cell line by qPCR (top) and Western blot (bottom); an unspecific band was used as a loading control. (B) E10.5 and E11.5 AGM cells were cultured and transduced with the same infective viral particles of shControl and shCdca7 (mix 1: shCdca7-1 + shCdca7-2; mix 2: shCdca7-1 + shCdca7-3). After 1 wk, the cells were analyzed by flow cytometry (CD45+cKit+: hematopoietic progenitors; CD45+cKit−: differentiated hematopoietic cells). One representative experiment is shown. (C) Mean of three independent experiments is plotted. (D) Cell cycle profile and apoptosis was determined in cells after 1 wk of culture. Graphs represent mean percentage of cells in each phase (left) and percentage of cells with Annexin V staining (right) from two independent experiments. No statistically significant differences were found. (E) After 1 wk in the hematopoietic culture, 3,000 cells were plated in a CFC culture. Bars represent the mean and SD of CFCs obtained from three independent cultures. Significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Cdca7 is required for in vivo definitive hematopoietic development in zebrafish
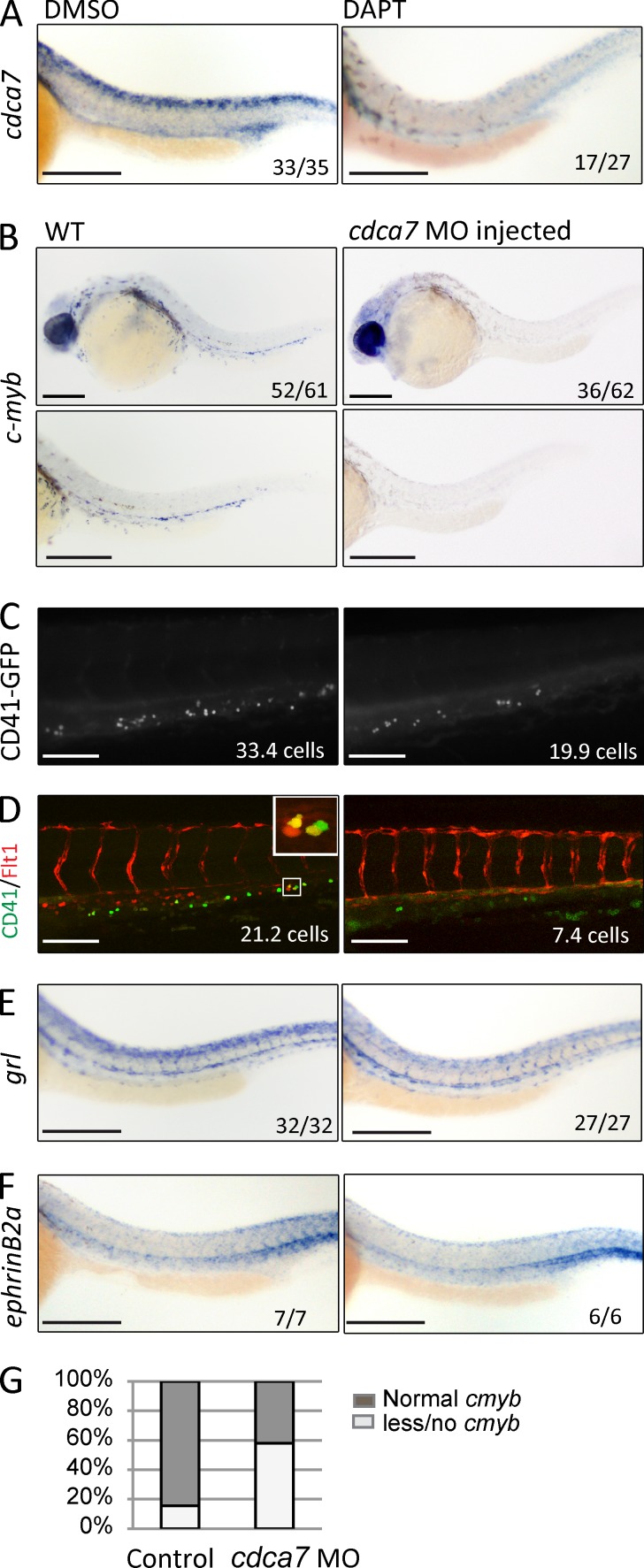
We first confirmed the expression pattern of cdca7 in the zebrafish embryos by ISH and found expression in the aorta of 94.3% of WT embryos (DMSO-treated control), whereas in vivo treatment with the Notch/γ-secretase inhibitor DAPT led to a severe reduction of cdca7 levels in 63% of treated embryos (n = 2, 33/35 DMSO vs. 10/27 DAPT showing normal cdca7 levels; Fig. 5 A; Thisse et al., 2001). These results indicate that cdca7 expression is also downstream of Notch in this system.
Figure 5.
HSC formation is impaired in cdca7 morphants. (A) cdca7 ISH of representative embryos treated at 6 hpf for 24 h with DMSO or 100 µM DAPT. (B) Representative embryos (top) and detailed images of AGM region (bottom) from embryos treated with control or cdca7 MOs. Hematopoietic cells were detected by cmyb ISH at 30 hpf or by CD41-GFP at 50 hpf (C). The number of CD41-GFP–positive cells is significantly reduced upon cdca7 MO injection (P = 0.01). (D) Representative images from CD41-GFP and Flt1-RFP embryos treated with control or cdca7 MOs. Number of double-positive cells is indicated. A detail of CD41-GFP+/Flt1-RFP+ HSCs is depicted in the inset. (E and F) The dorsal aorta integrity is revealed by gridlock (hey2) and EphrinB2a expression in both WT and cdca7 MO-injected embryos. The numbers in the panels indicate the number of embryos of the total pool that displayed phenotype or the number of positive cells. (G) The percentages of cmyb expression are indicated in the graph. Bars: (A, B, E, and F) 200 µm; (C and D) 100 µm.
Next, we tested the requirement of cdca7 during the HSC generation process in zebrafish. We took advantage of antisense cdca7 morpholino oligos (MOs) that had been previously tested (Ma et al., 2012; 2.3-fold reduction detected by qRT-PCR [not depicted]). By ISH, we detected cmyb expression (specifically labeling definitive HSCs and progenitors) in 84.4% of WT embryos compared with 41.9% of the embryos injected with cdca7 MOs (n = 3, 77 control embryos and 62 morphants analyzed; Fig. 5, B and G). Similar results were obtained with a second ATG MO against cdca7 (57.1% of treated embryos showed cmyb expression; not depicted). Moreover, the reduction in runx1 expression as detected by ISH indicated that cdca7 was required for specification of hemogenic endothelium (7/14 untreated embryos showed runx1 expression compared with 1/8 in the cdca7 MO-injected ones [not depicted]). Similarly, cdca7 MO treatment led to a reduction in the number of GFP-positive hematopoietic cells in the CD41-GFP line (Lin et al., 2005; 33.7 ± 3.6 in the untreated compared with 19.9 ± 3.0 cells in the MO-treated, P = 0.009; n = 1, 13, and 14 embryos, respectively; Fig. 5 C). The fact that CD41-GFP marks both HSCs and erythroid/myeloid progenitors (EMPs), with EMPs being independent of Notch signaling (Bertrand et al., 2010), is in agreement with the observation that the number of CD41-GFP–positive cells was only reduced by 33% in the MO-treated embryos. EMPs have a posterior blood island/venous origin (Lawson and Weinstein, 2002), whereas HSCs have an arterial AGM origin (Warga et al., 2009). Consequently, detection of Flt1-RFP, which is exclusively expressed in arterial cells (Bussmann et al., 2010), together with CD41 can distinguish between EMPs and HSCs (because RFP is very stable and HSCs remain RFP positive in the caudal hematopoietic tissue [CHT; Bertrand et al., 2008]). Quantification of the number of HSCs (CD41-GFP+/Flt1-RFP+) in the CHT demonstrated that control embryos contain 21.2 ± 3.3 GFP+/RFP+ compared with 7.4 ± 3.1 cells in the cdca7 MO-injected embryos (data from one experiment; six and nine embryos analyzed, respectively; P = 0.01; Fig. 5 D). Supporting the notion that cdca7 knockdown does not affect arterial development, we did not detect any significant alteration in the expression levels of the arterial genes gridlock and EphrinB2a by ISH and expression of Tg(Flt1-RFP) in our MO-treated embryos (Fig. 5, D–F). Together, our results demonstrate that CDCA7 is downstream of Notch/RBPj and contributes to the emergence of HSCs and maintenance of hematopoietic progenitors during embryonic development.
DISCUSSION
Our genome-wide analysis of RBPj-targeted genes during embryonic hematopoietic development provides a source of candidates to function as Notch mediators during HSC specification. Because it is still under debate which is the embryonic population of cells that requires Notch activity to give rise to HSCs, we have used pools of dissected AGMs from E11.5 embryos to search for RBPj-binding promoters. To further define the best candidate genes for Notch-dependent regulation in the AGM, we have focused on those that contain conserved adjacent RBPj-binding sites (Arnett et al., 2010) and showed Notch1 recruitment in their promoters. Furthermore, because Jag1-deficient embryos display specific hematopoietic defects (Robert-Moreno et al., 2008), we have selected genes whose expression depends on Jag1 in the AGM. In this work, we have focused on the Cdca7 gene, which scored positive to all of the aforementioned criteria, as a new Notch target gene involved in early hematopoietic development. In fact, we found that CDCA7 is required for the HSC and hematopoietic progenitor emergence in the zebrafish embryo.
Understanding the cellular and molecular mechanisms involved in HSC development is crucial for multiple therapeutic applications such as generation of HSCs from ESCs or induced pluripotent stem cells. This includes deciphering how endothelial-like cells acquire the hematopoietic fate and how a very restricted population achieves the stemness characteristics (LT-HSC; Taoudi et al., 2008) instead of just going through the hematopoietic pathway. We and others have previously shown that extra-embryonic yolk sac and embryonic stem–derived hematopoiesis do not require Notch activity (Schroeder et al., 2003; Robert-Moreno et al., 2007; Real et al., 2012), whereas it is essential to obtain transplantable definitive HSCs (Kumano et al., 2003). Here we found that Cdca7 is induced in a Notch-dependent manner during the initial stages of ESC differentiation into the hematopoietic lineage, but its expression is not maintained in the CD45+ mature population. Our results strongly suggest that sustained Notch signal and Cdca7 expression contribute to prevent maturation of hematopoietic progenitor/stem cells. In agreement with this possibility, we found that down-regulation of Cdca7 in AGM cells results in forced differentiation of the cKit+ progenitor population. In the ESC differentiation system, CDCA7 is temporarily activated in the CD34+CD45− HEPs but reduced in the CD45+ population, which could partially contribute to the engraftment inability of ESC-derived hematopoietic cells.
Cdca7 was previously identified as a downstream target of Myc and E2F transcription factors (Haggerty et al., 2003; Goto et al., 2006). CDCA7 can also interact with Myc in the nucleus and participate in Myc-dependent transformation events. It is proposed that CDCA7 participates in cell cycle progression, which may be relevant for its involvement in oncogenic processes (Whitfield et al., 2002). In this sense, CDCA7 is overexpressed in a variety of hematological tumors, predominantly in chronic myelogenous leukemia, but it has limited cell-transforming capacity both in vivo and in vitro (Osthus et al., 2005). However, we have not found any effect of CDCA7 on proliferation, nor cell death, in AGM hematopoietic cultures or in zebrafish hematopoietic cells (unpublished data). The precise physiological and biochemical function of CDCA7 remains obscure and requires further investigation. Here we show that Cdca7 is expressed in different HSC and progenitor subpopulations, including the LT-HSCs, and it is important for maintaining the undifferentiated phenotype. Moreover, our study shows that Cdca7 is downstream of Notch in hematopoietic cells in zebrafish, mouse, and human, indicating the high conservation of the Notch/RBPj/CDCA7 axis in hematopoietic development (Ciau-Uitz et al., 2014). Interestingly, Cdca7 expression was also enriched in the lgr5+ intestinal stem cells (van der Flier et al., 2009), suggesting a more general stemness-related function.
MATERIALS AND METHODS
Animals.
CD1, C57B6/J WT, and Jagged1−/− (Xue et al., 1999) strains were used. Animals were housed under pathogen-free conditions, and all procedures were approved by the Animal Care Committee of Parc de Recerca Biomèdica de Barcelona and Generalitat de Catalunya. Embryos were obtained from timed pregnant females and staged by somite counting: E10.5 (31–40 sp), E11.5 (43–48). The detection of the vaginal plug was designated as day 0.5. Mice and embryos were genotyped by PCR.
qRT-PCR.
Total RNA was extracted with a QIAGEN kit, and the RT-First Strand cDNA Synthesis kit (GE Healthcare) was used to produce cDNA. qRT-PCR was performed in the LightCycler480 system using SYBR Green I Master kit (Roche). Primers used are shown in Tables S2 and S3.
Promoter analysis and luciferase assays.
Cdca7 promoter was analyzed using Genomatix software. For luciferase assays, Cdca7 was generated cloning the region from −511 to 50 bp of Cdca7 TSS into the pGL3 vector (primers: forward, 5′-TGCTAGATAAGAGGTGCTATGCTC-3′; and reverse, 5′-GTGCTAGCACCCACCAACC-3′) and verified by sequencing. Two nucleotide (CC>AA) mutations were introduced by PCR at the −260 site alone or at both −260 and −217 core sites in the Cdca7 reporter. Luciferase reporter assays were performed in HEK-293T cells. Cells were seeded in 12-well plates at 5 × 104 cells/well. Cdca7p reporter, ICN1, Mam, dn-RBPj, or irrelevant DNA and CMV–β-Galactosidase plasmids were transfected in triplicate. Cells were transfected using polyethylenimine (Polysciences, Inc.). To reduce endogenous ICN1 levels, HEK-293T cells were treated with γ-secretase inhibitor DAPT (EMD Millipore) at 50 µM final concentration. Luciferase was measured 72 h after transfection according to the manufacturer’s instructions (Luciferase Assay System; Promega). Expression levels of transfected proteins were verified by Western blot.
ChIP assay and ChIP-on-chip.
Chromatin was obtained from a pool of 40 dissected AGMs at E11.5. ChIP was performed as previously described (Aguilera et al., 2004) with minor modifications. In brief, cross-linked chromatin from E10.5 or E11.5 dissected AGMs was sonicated for 10 min, medium power, 0.5 interval with a Bioruptor (Diagenode) and precipitated with anti-RBPj (Chu and Bresnick, 2004), anti-ICN1 (Abcam), and anti-Mam (Santa Cruz Biotechnology, Inc.). After cross-linkage reversal, DNA was used as a template for PCR or for array hybridization. Mouse promoter ChIP-on-chip microarray SET (Agilent Technologies) was used to identify RBPj targets. It covers 70,000 best identified gene regions with a −5.5- to 2.5-kb range and has, on average, 25 probes per gene with a mean probe to probe distance of 200 bp. The ChIP-on-chip was performed with dye swaps and one IgG control was brought along. Enrichment analysis was done by comparing the precipitation normalized dye swap signal with input control signal. Alternatively, qPCR was performed with SYBR Green I Master (Roche) in the LightCycler480 system. Primers used are in Table S4.
ChIP-on-chip analysis.
For calling the enriched peaks we used two independent algorithms, namely Chipper91 and Bioconductor92 package (version 2.7) iChIP (both iChIP1 and iChIP2; version 1.4.0). In the latter, background was corrected and Loess normalized data were used as input. For this purpose, the Bioconductor package Limma 93 was used. Multiple test-corrected (false discovery rate [FDR]; cutoff 0.05 for Chipper and 0.01 for iChIP) enriched probes were annotated to Ensembl (version 60) genes94. The RBPj target genes identified by both Chipper and iChIP were put in a highly confident RBPj targets dataset. Of this dataset, the region −5 to 5 kb from the TSS was scanned using the STORM program (version 9.4; 95) and Transfac RBPj position frequency matrix96 (using V$RBPJK_01, M01112) for putative in silico RBPj binding site determination. GO (Ashburner et al., 2000) functional and pathway (KEGG) enrichment analysis of genes was done by using GiTools97. Functional annotation of differentially target genes is based on GO (Ashburner et al., 2000), as extracted from Ensembl (Hubbard et al., 2007) and KEGG pathway databases (Kanehisa et al., 2008). Accordingly, all genes are classified into the ontology categories biological process (GOBP) and pathways when possible. We have taken only the GO/pathway categories that have at least 10 genes annotated. We used Gitools for enrichment analysis and heat map generation (Perez-Llamas and Lopez-Bigas, 2011). Resulting p-values were adjusted for multiple testing using the Benjamini and Hochberg method of FDR (Benjamini et al., 2001). All data have been deposited in the GEO database (accession no. GSE52094; included in Table S1).
Cell culture, viral particle production, and viral infection.
32D cells were cultured in Iscove’s and 10% FBS supplemented with 10% WeHi-IL3–conditioned media. Hematopoietic cells from disrupted AGMs were incubated in Iscove’s, 10% FBS, and 10% IL3– and 10% stem cell factor (SCF)–conditioned medium plus 0.2 µg/ml IL6 and 0.1 µg/ml flt3. Recombinant lentiviruses were produced by transient transfection of HEK-293T. In brief, subconfluent HEK-293T were cotransfected with 20 µg of transfer vector, 15 µg of packaging plasmid (psPAX2), and 6 µg of envelope plasmid (pMD2.G). After 3 d, supernatant was ultracentrifuged in a Beckman Coulter L-70 at 26,000 rpm for 2 h at 4°C and viral pellet resuspended in 100 ml PBS. Viruses were titrated in the 32D cell line, and the same amount of virus was used to infect the different conditions. Cultures were maintained for up to 12 d. Cells were harvested by pipetting and examined for hematopoietic development by flow cytometry.
Hematopoietic colony assay (CFC).
The dissected AGMs were digested in 0.12% collagenase (Sigma-Aldrich) in PBS supplemented with 10% FBS for 20 min at 37°C. AGM disrupted cells were cultured 7 d in Iscove’s, 10% FBS, and 10% IL3– and 10% SCF–conditioned medium plus 0.2 µg/ml IL6 and 0.1 µg/ml flt3. Cells were infected with the same amount of infective particles (shControl or shCdca7), and puromycin was added the following day. After 1 wk, cells were analyzed by FACS, and 3,000 cells from each condition were plated in duplicates in M-3434 semisolid medium (STEMCELL Technologies) to perform a CFC assay. After 7 d, the presence of hematopoietic colonies was scored under the microscope.
Flow cytometry analysis and sorting.
The AGM region was dissected from E10.5 embryos and dissociated in 0.12% collagenase (Sigma-Aldrich) in PBS supplemented with 10% FBS for 20 min at 37°C. The cells were analyzed by flow cytometry in a FACS Fortessa (BD). FlowJo software was used to analyze the results. Dead cells were excluded by DAPI. The antibodies CD31-PE, CD45-PECy7, and CD117-APC were purchased from BD. After staining for CD45, cells were fixed (Invitrogen) for cell cycle analysis, washed with PBS and 10% FBS, and stained with anti–Ki67-APC (BD) in permeabilizing buffer (Invitrogen) for 20 min at room temperature. Then, cells were resuspened in PBS containing 2 µg/ml DAPI (Invitrogen). Annexin V staining was performed as indicated by the apoptosis detection kit (eBioscience), and cells were analyzed in binding buffer containing 5 µg/ml DAPI (Invitrogen). Cells were analyzed by flow cytometry.
ISH and immunofluorescence.
For ISH, precisely timed embryos were fixed overnight at 4°C in 4% paraformaldehyde and subsequently embedded in OCT (Tissue-Tek). Embryos were sectioned in a Leica RM2135 cryostat at 16 µm. Digoxigenin (DIG)-labeled probes were generated from a pGEMT easy plasmid (Promega) containing Cdca7 cDNA (75/1,220). 1.1 kb of Cdca7 transcript was cloned using the following: forward, 5′-AAAGAATTCGAGGCTCGCCGCGCGC-3′; and reverse, 5′-AAACTCGAGCTACGCTTGCATTTCAAATTC-3′. Images were acquired with an Olympus BX-60. For immunostaining, embryos at E10.5 and E11.5 were fixed overnight at 4°C in 37% formaldehyde and subsequently dehydrated with ethanol and embedded in paraffin. Anti-CDCA7 antibody was developed by AVIVA (ARP66756). Images were acquired in confocal microscope (Leica SP5). Representative images were edited on Adobe Photoshop CS4 software.
hESC hematopoietic cell differentiation and cell sorting.
hESC-OP9 co-cultures were performed as previously described (Vodyanik and Slukvin, 2007) with some modifications (Real et al., 2012). In brief, OP9 stroma was prepared in gelatin-coated 10-cm dishes in α-MEM basal medium supplemented with 20% nonheat-inactivated FBS. The hESC lines AND1 and H9 grown in Matrigel-coated flasks were prepared as a suspension of small aggregates using Collagenase IV treatment followed by gentle scraping in differentiation medium (DM; α-MEM basal medium, 10% nonheat-inactivated FBS, 100 mM monothioglycerol, and 50 mg/ml ascorbic acid) and plated on top of the 8-d OP9 stroma in 10 ml DM. The next day, media were replaced by 20 ml DM to remove unattached cells; where indicated, we added 200 nM Compound E (CompE; EMD Millipore) to block Notch activation. From day 3 of co-culture, a half-volume media change was performed every other day. At days 8 (for HEP purification) and 15 of co-culture (for CD45+ purification), cells were treated for 1 h with collagenase IV, followed by 20 min with Tryple (Gibco). Single cell suspensions were stained with anti–mouse CD29-FITC, anti–human CD31-PE, CD34-PE-Cy7, and CD45-APC antibodies, and human HEPs and CD45+ cells were purified by cell sorting as described above within the CD29− human population.
To allow hEB formation, undifferentiated hESCs were treated with collagenase IV and scraped off the Matrigel. These hESC clumps were transferred to low-attachment plates (Corning) and incubated overnight in DM (KO-DMEM), 20% nonheat-inactivated FBS, 1% nonessential amino acids, 1 mmol/l l-glutamine, and 0.1 mmol/l β-mercaptoethanol. From day 1 the media was changed every 3 d to the same DM supplemented with hematopoietic cytokines: 300 ng/ml SCF, 300 ng/ml Flt3L, 10 ng/ml IL-3, 10 ng/ml IL-6, 50 ng/ml G-CSF, and 25 ng/ml bone morphogenetic protein 4 (BMP-4; Ramos-Mejia et al., 2012). At day 15 of differentiation, hEBs were dissociated using collagenase B (Roche) for 2 h at 37°C, followed by 10-min incubation at 37°C with enzyme-free cell dissociation buffer (Invitrogen). The dissociated cells were stained as indicated above. Work with hESCs has been approved by the “Comisión Nacional de Control y Garantías” del Instituto de Salud Carlos III.
Zebrafish maintenance, morpholinos, microinjection and ISH.
Embryos and adult fish were raised and maintained under standard laboratory conditions in the Erasmus Animal Facility (EDC). For the cdca7 probe, cdca7 full length was cloned from cDNA using the primers forward, 5′-TATTATGAATTCATGAACCTGCGGAGCTAC-3′; and reverse, 5′-TATTATTCTAGATTATTCACTTTCCTCCAG-3′. Subsequently, the cdca7 antisense probe was made using 500 bp at the 3′ end of the gene using a digest of the natural Bgl1 restriction site in cdca7. DAPT treatment was performed according to Geling et al. (2002). Dechorionated embryos were treated at 6 h postfertilization (hpf) for 24 h in E3 containing either DMSO or DAPT. A 10-mM stock (in DMSO) was diluted in E3 medium to a final concentration of 100 µM. Embryos were fixed in 4% paraformaldehyde containing sucrose for ISH.
An antisense splice blocking MO against cdca7 (3′-TACTGTAGATACAAACCATTTCTGT-5′), an ATG blocking MO against cdca7 (3′-AGCTCCTGTAGCTCCGCAGGTTCAT-5′), and p53 have been previously described (Ma et al., 2012) and were purchased from Gene Tools. MOs (4 ng) were injected alone or in combination with p53 MO into one-cell-stage zebrafish embryos. MO knockdown was tested using the following primer sets: cdca7 forward, 3′-CAGACGAGAAGCTCCACTCA-5′; cdca7 reverse, 3′-TAGGCTCAGGCTTCACTGGT-5′; elfa qRT-PCR control: elfa forward, 3′-CTTCTCAGGCTGACTGTGC-5′; and elfa reverse, 3′-CCGCTAGCATTACCCTCC-5′. WISH for zebrafish embryos was performed at 30 hpf as described previously (Patterson et al., 2005). The CD41-GFP and CD41-GFP/Flt1-RFP imaging was performed at 50 hpf and the area imaged was the CHT. cmyb, gridlock (grl), and ephrinB2a probes (Thompson et al., 1998) and CD41-GFP (Lin et al., 2005) were used as markers for HSCs in zebrafish. Flt1-RFP was used as a marker for arterial cells (Bussmann et al., 2010). CD41-GFP and Flt1-RFP transgenic zebrafish were kept in a homozygous state. Significance was determined by the Student’s t test.
Online supplemental material.
Fig. S1 shows that Hes1 predicted RBPj-binding sites. Fig. S2 shows Cdca7 is expressed in human and mouse hematopoietic progenitors. Table S1, included as a separate PDF file, shows Chipper- and iChip-identified RBPj target genes. Tables S2 and S3 show mouse and human expression RT-PCR primers, respectively. Table S4 shows promoter mouse primers. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131857/DC1.
Supplementary Material
Acknowledgments
We thank E.H. Bresnick (University of Wisconsin, Madison, WI) for the anti-RBPj antibody; Leonard Zon and Michelle I. Lin (Harvard Medical School, Boston, MA) for sharing unpublished ChIP-seq data; the microarray facility, animal facility, and flow cytometry facility at Parc de Recerca Biomèdica de Barcelona for technical assistance; Alba Placin and Vera Horvath for technical contribution to the project; Miguel Campanero (Instituto de Investigaciones Biomédicas, Madrid, Spain) and Pedro Real (GENIO, Seville, Spain) for support; and the members of the Bigas laboratory for helpful discussions.
J. Guiu was a recipient of Formación Personal Investigador (FPI) BES-2008-005708. This research was funded by grants from the Ministerio de Economía y Competitividad, FEDER (SAF2007-60080, PLE2009-0111, SAF2010-15450, and SAF2013-40922R), Red Temática de Investigación Cooperativa en Cáncer (RTICC; RD06/0020/0098 and RD12/0036/0054), Association for International Cancer Research (AICR; 13-0064), Agència de Gestió d’Ajuds Universitaris i de Recerca (AGAUR; 2009SGR-23, CONES2010-0006) to A. Bigas, the Spanish Association against Cancer Research (AECC) to A. Bigas and P. Menendez, Fondo de Investigación Sanitaria (FIS; PI10/00449) to P. Menendez, and ZonMW TOP (40-00812-98-11068) to E. Dzierzak and E. De Pater.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AGM
- aorta-gonad-mesonephros
- ChIP
- chromatin immunoprecipitation
- CHT
- caudal hematopoietic tissue
- EB
- embryoid body
- EMP
- erythroid/myeloid progenitor
- ESC
- embryonic stem cell
- HEP
- hematoendothelial precursor
- hpf
- hour postfertilization
- HSC
- hematopoietic stem cell
- ISH
- in situ hybridization
- MO
- morpholino oligo
- SCF
- stem cell factor
- TSS
- transcriptional start site
References
- Aguilera C., Hoya-Arias R., Haegeman G., Espinosa L., and Bigas A.. 2004. Recruitment of IκBα to the hes1 promoter is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA. 101:16537–16542 10.1073/pnas.0404429101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett K.L., Hass M., McArthur D.G., Ilagan M.X., Aster J.C., Kopan R., and Blacklow S.C.. 2010. Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat. Struct. Mol. Biol. 17:1312–1317 10.1038/nsmb.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. . The Gene Ontology Consortium. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Drai D., Elmer G., Kafkafi N., and Golani I.. 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125:279–284 10.1016/S0166-4328(01)00297-2 [DOI] [PubMed] [Google Scholar]
- Bertrand J.Y., Kim A.D., Teng S., and Traver D.. 2008. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 135:1853–1862 10.1242/dev.015297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Cisson J.L., Stachura D.L., and Traver D.. 2010. Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood. 115:2777–2783 10.1182/blood-2009-09-244590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigas A., and Espinosa L.. 2012. Hematopoietic stem cells: to be or Notch to be. Blood. 119:3226–3235 10.1182/blood-2011-10-355826 [DOI] [PubMed] [Google Scholar]
- Bueno C., Ayllón V., Montes R., Navarro-Montero O., Ramos-Mejia V., Real P.J., Romero-Moya D., Araúzo-Bravo M.J., and Menendez P.. 2013. FLT3 activation cooperates with MLL-AF4 fusion protein to abrogate the hematopoietic specification of human ESCs. Blood. 121:3867–3878 10.1182/blood-2012-11-470146 [DOI] [PubMed] [Google Scholar]
- Burns C.E., Traver D., Mayhall E., Shepard J.L., and Zon L.I.. 2005. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19:2331–2342 10.1101/gad.1337005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann J., Bos F.L., Urasaki A., Kawakami K., Duckers H.J., and Schulte-Merker S.. 2010. Arteries provide essential guidance cues for lymphatic endothelial cells in the zebrafish trunk. Development. 137:2653–2657 10.1242/dev.048207 [DOI] [PubMed] [Google Scholar]
- Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., and Speck N.A.. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 457:887–891 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., and Bresnick E.H.. 2004. Evidence that C promoter-binding factor 1 binding is required for Notch-1-mediated repression of activator protein-1. J. Biol. Chem. 279:12337–12345 10.1074/jbc.M311510200 [DOI] [PubMed] [Google Scholar]
- Ciau-Uitz A., Monteiro R., Kirmizitas A., and Patient R.. 2014. Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp. Hematol. 42:669–683 10.1016/j.exphem.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Duarte A., Hirashima M., Benedito R., Trindade A., Diniz P., Bekman E., Costa L., Henrique D., and Rossant J.. 2004. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 18:2474–2478 10.1101/gad.1239004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geling A., Steiner H., Willem M., Bally-Cuif L., and Haass C.. 2002. A γ-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 3:688–694 10.1093/embo-reports/kvf124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Hayashi R., Muramatsu T., Ogawa H., Eguchi I., Oshida Y., Ohtani K., and Yoshida K.. 2006. JPO1/CDCA7, a novel transcription factor E2F1-induced protein, possesses intrinsic transcriptional regulator activity. Biochim. Biophys. Acta. 1759:60–68 10.1016/j.bbaexp.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Guiu J., Shimizu R., D’Altri T., Fraser S.T., Hatakeyama J., Bresnick E.H., Kageyama R., Dzierzak E., Yamamoto M., Espinosa L., and Bigas A.. 2013. Hes repressors are essential regulators of hematopoietic stem cell development downstream of Notch signaling. J. Exp. Med. 210:71–84 10.1084/jem.20120993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty T.J., Zeller K.I., Osthus R.C., Wonsey D.R., and Dang C.V.. 2003. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc. Natl. Acad. Sci. USA. 100:5313–5318 10.1073/pnas.0931346100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., et al. . 2007. Ensembl 2007. Nucleic Acids Res. 35:D610–D617 10.1093/nar/gkl996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovs A., Rybtsov S., Welch L., Anderson R.A., Turner M.L., and Medvinsky A.. 2011. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J. Exp. Med. 208:2417–2427 10.1084/jem.20111688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., and Yamanishi Y.. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36:D480–D484 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konuma T., Nakamura S., Miyagi S., Negishi M., Chiba T., Oguro H., Yuan J., Mochizuki-Kashio M., Ichikawa H., Miyoshi H., et al. . 2011. Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp. Hematol. 39:697–709: e5 10.1016/j.exphem.2011.03.008 [DOI] [PubMed] [Google Scholar]
- Krebs L.T., Shutter J.R., Tanigaki K., Honjo T., Stark K.L., and Gridley T.. 2004. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 18:2469–2473 10.1101/gad.1239204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejcí A., and Bray S.. 2007. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 21:1322–1327 10.1101/gad.424607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K., Chiba S., Kunisato A., Sata M., Saito T., Nakagami-Yamaguchi E., Yamaguchi T., Masuda S., Shimizu K., Takahashi T., et al. . 2003. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 18:699–711 10.1016/S1074-7613(03)00117-1 [DOI] [PubMed] [Google Scholar]
- Lawson N.D., and Weinstein B.M.. 2002. Arteries and veins: making a difference with zebrafish. Nat. Rev. Genet. 3:674–682 10.1038/nrg888 [DOI] [PubMed] [Google Scholar]
- Lee J.B., Werbowetski-Ogilvie T.E., Lee J.H., McIntyre B.A., Schnerch A., Hong S.H., Park I.H., Daley G.Q., Bernstein I.D., and Bhatia M.. 2013. Notch-HES1 signaling axis controls hemato-endothelial fate decisions of human embryonic and induced pluripotent stem cells. Blood. 122:1162–1173 10.1182/blood-2012-12-471649 [DOI] [PubMed] [Google Scholar]
- Lin H.F., Traver D., Zhu H., Dooley K., Paw B.H., Zon L.I., and Handin R.I.. 2005. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 106:3803–3810 10.1182/blood-2005-01-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Wang L., Wang S., Gao Y., Wei Y., and Liu F.. 2012. Foxn1 maintains thymic epithelial cells to support T-cell development via mcm2 in zebrafish. Proc. Natl. Acad. Sci. USA. 109:21040–21045 10.1073/pnas.1217021110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal L., Banerjee U., and Hartenstein V.. 2004. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat. Genet. 36:1019–1023 10.1038/ng1404 [DOI] [PubMed] [Google Scholar]
- McKinney-Freeman S., Cahan P., Li H., Lacadie S.A., Huang H.T., Curran M., Loewer S., Naveiras O., Kathrein K.L., Konantz M., et al. . 2012. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 11:701–714 10.1016/j.stem.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthus R.C., Karim B., Prescott J.E., Smith B.D., McDevitt M., Huso D.L., and Dang C.V.. 2005. The Myc target gene JPO1/CDCA7 is frequently overexpressed in human tumors and has limited transforming activity in vivo. Cancer Res. 65:5620–5627 10.1158/0008-5472.CAN-05-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson L.J., Gering M., and Patient R.. 2005. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 105:3502–3511 10.1182/blood-2004-09-3547 [DOI] [PubMed] [Google Scholar]
- Perez-Llamas C., and Lopez-Bigas N.. 2011. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE. 6:e19541 10.1371/journal.pone.0019541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii S., Kloss C.C., Butler J.M., Ginsberg M., Gars E., Lis R., Zhan Q., Josipovic P., Ding B.S., Xiang J., et al. . 2013. Human ESC-derived hemogenic endothelial cells undergo distinct waves of endothelial to hematopoietic transition. Blood. 121:770–780 10.1182/blood-2012-07-444208 [DOI] [PubMed] [Google Scholar]
- Ramos-Mejia V., Bueno C., Roldan M., Sanchez L., Ligero G., Menendez P., and Martin M.. 2012. The adaptation of human embryonic stem cells to different feeder-free culture conditions is accompanied by a mitochondrial response. Stem Cells Dev. 21:1145–1155 10.1089/scd.2011.0248 [DOI] [PubMed] [Google Scholar]
- Real P.J., Ligero G., Ayllon V., Ramos-Mejia V., Bueno C., Gutierrez-Aranda I., Navarro-Montero O., Lako M., and Menendez P.. 2012. SCL/TAL1 regulates hematopoietic specification from human embryonic stem cells. Mol. Ther. 20:1443–1453 10.1038/mt.2012.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Moreno A., Espinosa L., de la Pompa J.L., and Bigas A.. 2005. RBPjκ-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 132:1117–1126 10.1242/dev.01660 [DOI] [PubMed] [Google Scholar]
- Robert-Moreno A., Espinosa L., Sanchez M.J., de la Pompa J.L., and Bigas A.. 2007. The notch pathway positively regulates programmed cell death during erythroid differentiation. Leukemia. 21:1496–1503 10.1038/sj.leu.2404705 [DOI] [PubMed] [Google Scholar]
- Robert-Moreno A., Guiu J., Ruiz-Herguido C., López M.E., Inglés-Esteve J., Riera L., Tipping A., Enver T., Dzierzak E., Gridley T., et al. . 2008. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 27:1886–1895 10.1038/emboj.2008.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herguido C., Guiu J., D’Altri T., Inglés-Esteve J., Dzierzak E., Espinosa L., and Bigas A.. 2012. Hematopoietic stem cell development requires transient Wnt/β-catenin activity. J. Exp. Med. 209:1457–1468 10.1084/jem.20120225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T., Fraser S.T., Ogawa M., Nishikawa S., Oka C., Bornkamm G.W., Nishikawa S., Honjo T., and Just U.. 2003. Recombination signal sequence-binding protein Jκ alters mesodermal cell fate decisions by suppressing cardiomyogenesis. Proc. Natl. Acad. Sci. USA. 100:4018–4023 10.1073/pnas.0438008100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S., Gonneau C., Moore K., Sheridan J.M., Blackburn C.C., Taylor E., and Medvinsky A.. 2008. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell. 3:99–108 10.1016/j.stem.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Terriente-Felix A., Li J., Collins S., Mulligan A., Reekie I., Bernard F., Krejci A., and Bray S.. 2013. Notch cooperates with Lozenge/Runx to lock haemocytes into a differentiation programme. Development. 140:926–937 10.1242/dev.086785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Pflumio S., Fürthauer M., Loppin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X.Q., and Thisse C.. 2001. Expression of the zebrafish genome during embryogenesis. ZFIN, University of Oregon, Eugene, OR: Available at: http://zfin.org/ZDB-PUB-010810-1 (accessed March 11, 2014). [Google Scholar]
- Thompson M.A., Ransom D.G., Pratt S.J., MacLennan H., Kieran M.W., Detrich H.W. III, Vail B., Huber T.L., Paw B., Brownlie A.J., et al. . 1998. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev. Biol. 197:248–269 10.1006/dbio.1998.8887 [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., van Gijn M.E., Hatzis P., Kujala P., Haegebarth A., Stange D.E., Begthel H., van den Born M., Guryev V., Oving I., et al. . 2009. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 136:903–912 10.1016/j.cell.2009.01.031 [DOI] [PubMed] [Google Scholar]
- Vodyanik M.A., and Slukvin I.I.. 2007. Hematoendothelial differentiation of human embryonic stem cells. Curr. Protoc. Cell Biol. Chapter 23:Unit 23.6. [DOI] [PubMed] [Google Scholar]
- Wang L., Li L., Shojaei F., Levac K., Cerdan C., Menendez P., Martin T., Rouleau A., and Bhatia M.. 2004. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 21:31–41 10.1016/j.immuni.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Warga R.M., Kane D.A., and Ho R.K.. 2009. Fate mapping embryonic blood in zebrafish: multi- and unipotential lineages are segregated at gastrulation. Dev. Cell. 16:744–755 10.1016/j.devcel.2009.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., and Botstein D.. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13:1977–2000 10.1091/mbc.02-02-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Gao X., Lindsell C.E., Norton C.R., Chang B., Hicks C., Gendron-Maguire M., Rand E.B., Weinmaster G., and Gridley T.. 1999. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 8:723–730 10.1093/hmg/8.5.723 [DOI] [PubMed] [Google Scholar]
- Yokomizo T., and Dzierzak E.. 2010. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 137:3651–3661 10.1242/dev.051094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L.R., Lin F.J., Lee C.T., DeMayo F.J., Tsai M.J., and Tsai S.Y.. 2005. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 435:98–104 10.1038/nature03511 [DOI] [PubMed] [Google Scholar]
- Yu X., Zou J., Ye Z., Hammond H., Chen G., Tokunaga A., Mali P., Li Y.M., Civin C., Gaiano N., and Cheng L.. 2008. Notch signaling activation in human embryonic stem cells is required for embryonic, but not trophoblastic, lineage commitment. Cell Stem Cell. 2:461–471 10.1016/j.stem.2008.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein A.C., Hofmann J.J., Lynch M., French W.J., Turlo K.A., Yang Y., Becker M.S., Zanetta L., Dejana E., Gasson J.C., et al. . 2008. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 3:625–636 10.1016/j.stem.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.