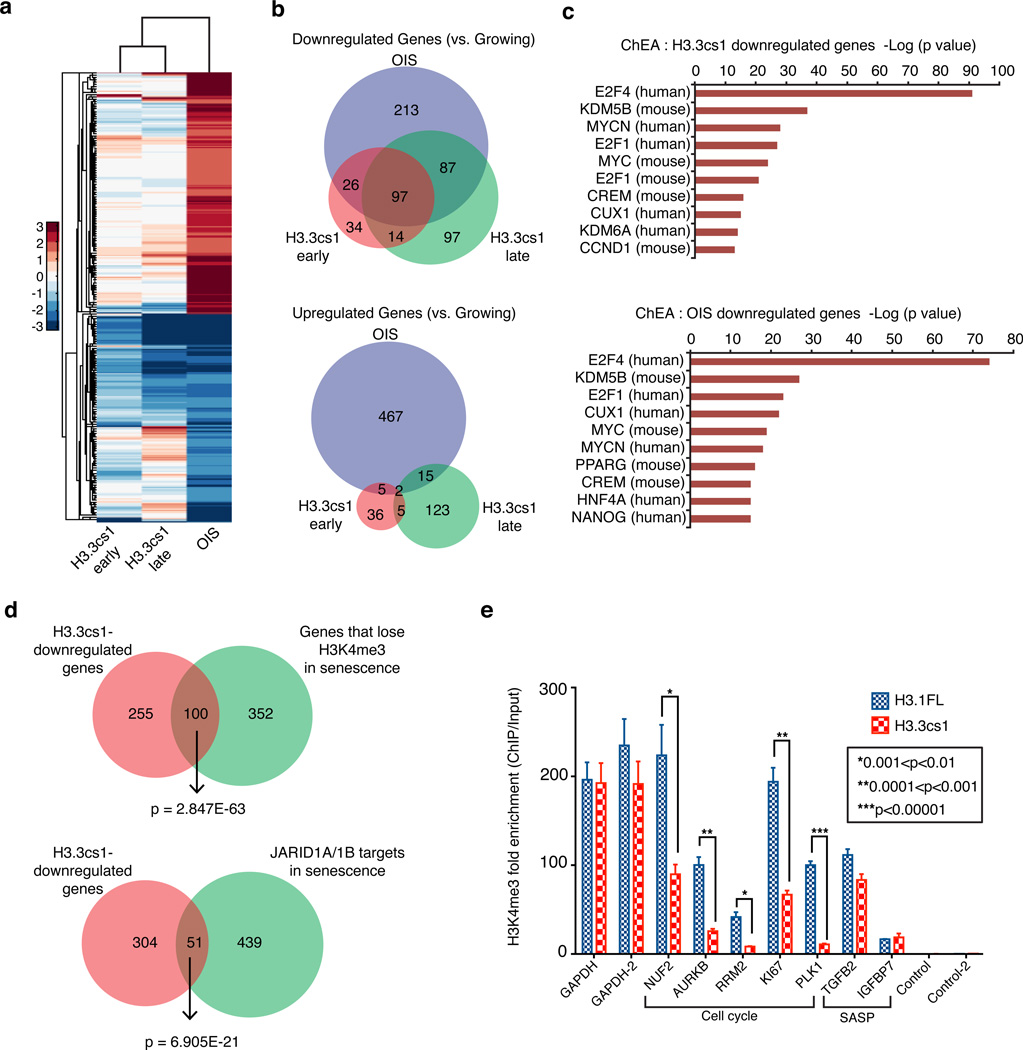
Figure 5. H3.3cs1-induced senescence is mediated by decreased transcriptional output of cell cycle genes and reduced H3K4me3.
(a) Heat map showing gene expression changes of 914 genes as determined by RNA-seq in H3.3cs1-induced senescence (early and late time points) relative to those that are altered in OIS (vs. growing). Note the overlap of downregulated genes between H3.3cs1-induced senescence and OIS. (b) (Top) Venn diagram of genes downregulated in OIS, H3.3cs1 (early) and H3.3cs1 (late) as compared to growing IMR90s. Fisher exact test; p= 2.418E-117 OIS vs. H3.3cs1 (early); p=3.477E-162 OIS vs. H3.3cs1 (late); p=2.404E-117 H3.3cs1 (early) vs. H3.3cs1 (late). (Bottom) Venn diagram of genes upregulated in OIS, H3.3cs1 (early) and H3.3cs1 (late) as compared to growing IMR90s. Fisher exact test; p=3.414E-4 OIS vs. H3.3cs1 (early); p= 2.883E-7 OIS vs. H3.3cs1 (late); p=1.783E-7 H3.3cs1 (early) vs. H3.3cs1 (late). (c) ChIP Enrichment Analysis (ChEA) of H3.3cs1-induced senescence (top) and OIS (bottom) downregulated genes. Note that E2F family members and KDM5B (JARID1B) gene targets are enriched in both. The −log of the p-value associated with each term is plotted. (d) Venn diagrams illustrating the overlap between H3.3cs1 downregulated genes and genes that lose H3K4me3 in senescence (left) and JARID1A/1B-regulated genes in senescence15. Fisher exact test of p values indicated in the diagram. (e) ChIP-qPCR for H3K4me3 at cell cycle genes and SASP-related genes in IMR90s expressing H3.1FL (blue) or H3.3cs1 (red). Two regions near the GAPDH promoter used as positive controls; intergenic regions used as negative controls. Fold enrichment of H3K4me3 ChIP/ Input is plotted. Mean ± s.e.m; two-tailed paired sample t-test; p-values as indicated.