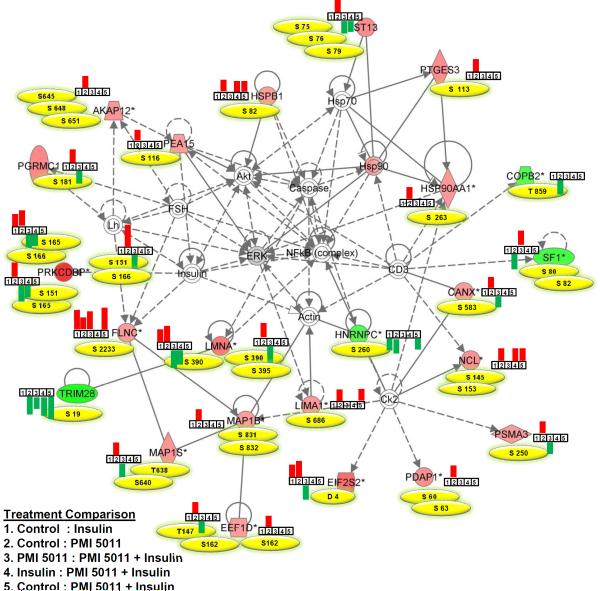
Figure 2.
Top connectivity network with p-value 10−65 showing changes in expression of phosphopeptides involved in NFκB network in basal and insulin stimulated states with and without PMI 5011 treatment. Proteins labeled with filled red or green colors were identified to contain differentially regulated phosphopeptides in PMI 5011 treated baseline control cultures. All other proteins are imported from the IPA knowledge base. Phosphorylation sites identified in each protein in the network are listed in yellow, and bar graphs show whether a phosphopeptide was up or down-regulated based on a specific treatment comparison. Up-regulated and down-regulated peptides are presented in red and green, respectively. A line indicates interactions, and the dotted lines indicate an inferred or indirect interaction. The abbreviations are: AKAP12: A kinase (PRKA) anchor protein 12, CANX: calnexin, COBP2: coatomer subunit beta, EEF1D: eukaryotic translation elongation factor 1 delta, EIF2S2: eukaryotic translation initiation factor 2 subunit 2, FLNC: filamin C, HNRNPC: heterogeneous nuclear ribonucleoprotein C, HSP90AA1: heat shock protein 90kDa alpha, HSPB1: heat shock 27kDa protein 1, LIMA1: LIM domain and actin binding 1, LMNA: lamin A/C, MAP1B: microtubule-associated protein 1B, MAP1S: microtubule-associated protein 1S, NCL: nucleolin, PDAP1: PDGFA associated protein 1, PEA15: phosphoprotein enriched in astrocytes 15, PGRMC1: progesterone receptor membrane component 1, PRKCDBP: protein kinase C, delta binding protein (Cavin-3), PSMA3: proteasome subunit alpha 3, PTGES3: predicted pseudogene 9769, SF1: splicing factor 1, ST13: Hsp70 interacting protein, TRIM28: tripartite motif containing 28.