Figure 4.
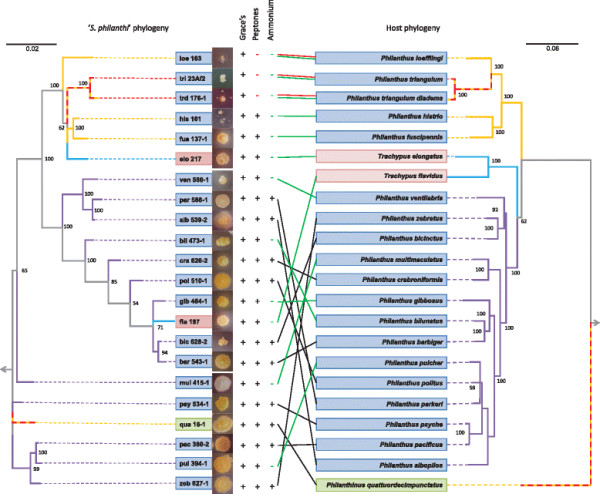
Phylogeny of ‘S. philanthus’ biovars in respect to their morphology, nutritional requirements and host phylogeny. The phylogeny of bacterial symbionts was reconstructed using nearly complete 16S rRNA genes, as well as gyrA and gyrB gene fragments (566 and 660 bp in length, respectively). The host phylogeny was obtained from [28]. Colored boxes around host and symbiont names denote host genera (green, Philanthinus; blue, Philanthus; red, Trachypus). Values at the nodes of the phylogenetic trees indicate Bayesian posterior probabilities. Geographic distribution of beewolf taxa and the origin of isolated symbionts are indicated by branches of different colours on phylogenetic trees: Africa (yellow), mixed African/Eurasian distribution (dashed yellow/red line), North America (purple) and South America (blue). Metabolic capabilities are indicated next to representative pictures of symbiont growth in vitro: Plus or minus indicate the ability (+) or inability (−) to grow on the corresponding media. In order to visualize the phylogenetic placement of symbionts and highlight their metabolic capabilities, symbiont strains were connected to their respective hosts with colored lines: Red lines correspond to strains unable to grow on medium with peptones; green lines correspond to strains unable to grow on ammonium as the only source of nitrogen.
