Figure 2.
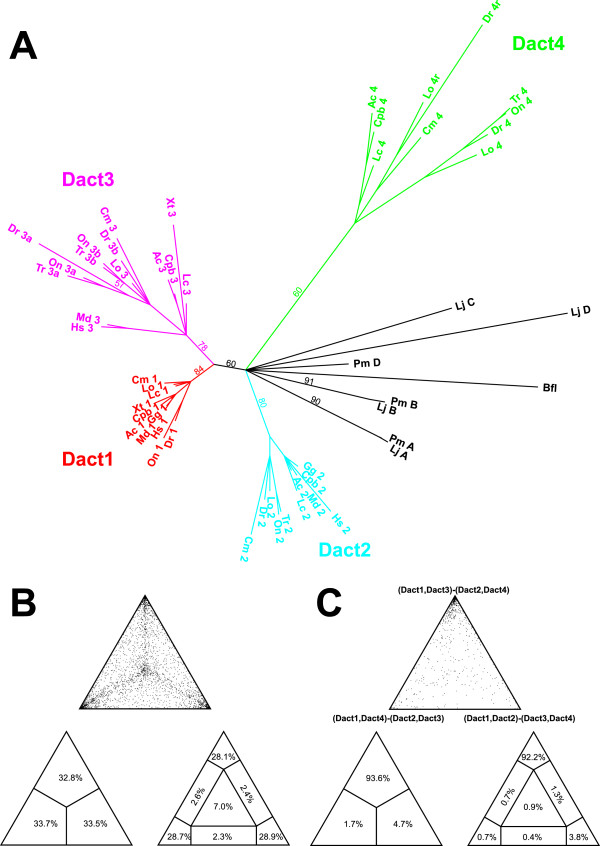
Phylogenetic analysis of Dact proteins. Reconstruction of the phylogenetic tree of Dact proteins and likelihood mapping by quartet puzzling using Tree-Puzzle. (A) Unrooted phylogenetic tree of Dact protein sequences from humans (Hs), opossum (Md), chicken (Gg), Anole lizard (Ac), the Western painted turtle (Cpb), Xenopus tropicalis (Xt), Latimeria (Lc), the spotted gar (Lo), zebrafish (Dr), Fugu (Tr), Tilapia (On), elephant shark (Cm), sea lamprey (Pm), Japanese lamprey (Lj), and Branchiostoma floridae (Bfl). The tree was created using the JTT model with accurate parameter estimation and using 100,000 puzzling steps. Likelihood values are indicated for branch points separating major groups. Sequences are annotated using the abbreviation for the species, followed by the Dact ortholog number. Note that the gnathostome Dact1 (red branches) and Dact3 sequences (pink branches) formed a metagroup. Dact2 (turquoise branches) and Dact4 sequences (green branches) each formed distinct groups. They emerge from a star-like node together with the four cyclostomes dact proteins and the Branchiostoma dact sequence, indicating the ambiguity of the tree topology for this part of the tree. (B) Likelihood mapping of the Dact protein sequences used for the phylogenetic tree reconstruction, based on 10,000 random quartets. 85.7% of quartets were fully resolved, indicating overall tree-like character. (C) Likelihood mapping of the Dact1, Dact2, Dact3 and Dact4 clusters, based on 10,000 random quartets. 92.2% of quartets support the Dact1/3 versus Dact2/4 subdivision.
