Figure 4.
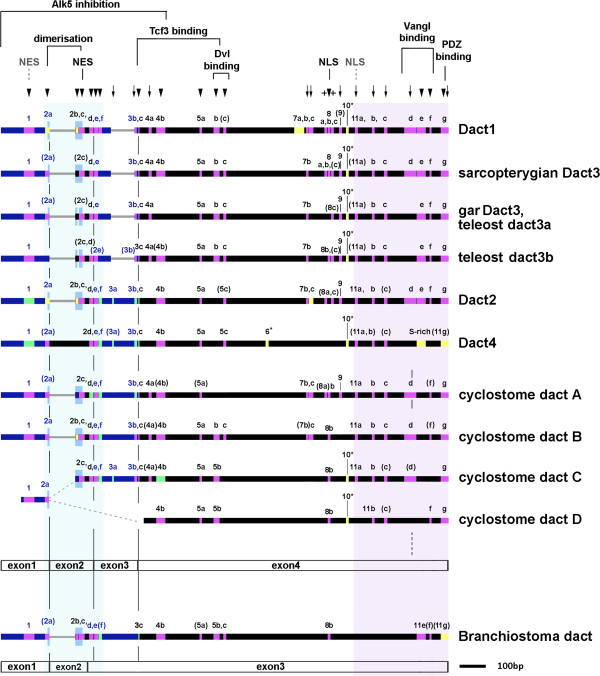
Conserved Dact protein motifs. Graphical display of the gapped Dact protein alignments (thick horizontal lines); large sequence stretches missing in a particular Dact are shown by thin grey lines. Purple: shared protein motifs;red: shared Dact1/3 motif variations; green: shared Dact2/4 motif variations in green; yellow: motifs typical for a particular Dact ortholog; turquoise: in motif 11 g, the cyclostome dactA and dactB proteins share amino acids that are specific for either Dact2 or Dact1/3. The leucine zipper is marked by mid-blue, higher boxes. The lengths of motifs are according to scale. The light blue and lilac background shading indicates the most conserved areas of Dact proteins. Motifs are numbered according to their position in the Dact alignment; linked motifs are marked by letters, partial versions of a motif are in brackets. Known roles of motifs or sequence stretches are indicated at the top, predicted roles are marked in grey and with dotted lines. Exon boundaries are indicated below a set of sequences; note that the exon2-3-4 boundaries are different in vertebrates and Branchiostoma, and that the cyclostome dactA features a fourth intron within motif 11d. In the cyclostomes, a genomic fragment carrying a recognizable dact exon1 sequence was not linked to the fragments carrying the dactC or dactD sequences, hence the exon1 carrying fragment may belong to either (dotted grey lines). Importantly, some motifs such as the leucine zipper are present already in the lancelet and hence, constitute the original repertoire of dacts (marked by arrowheads). Other motifs arose in the vertebrate (arrows) or, subsequently, in the gnathostome lineage (crosses). Gnathostome Dact orthologs have a unique composition of motifs. However, motifs are the most similar in Dact1/3 and Dact2/4, respectively. Cyclostome dact proteins resemble a mix of Dact1/3-typical sequences, Dact2/4 sequences, and unique sequences.
