Figure 6.
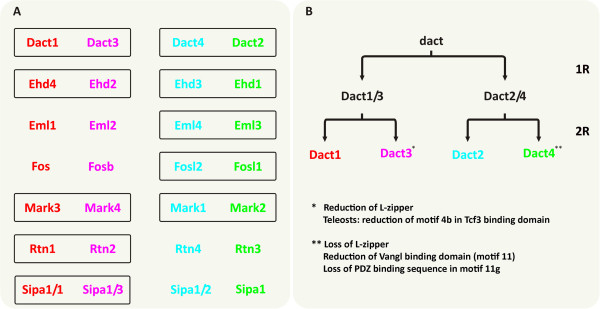
Summary of the phylogenetic analysis of gnathostome Dact and Dact associated genes. (A) Grouping of gnathostome Dact and Dact associated genes as suggested by the phylogenetic analysis of the cognate protein sequences. Genes genomically colocalizing with a particular Dact gene are depicted in the same color as the associated Dact gene. Black boxes link genes that form a well-supported metagroup in the corresponding phylogenetic tree (Additional file 7). Note that in all cases, Dact1/3 and/or Dact2/4 associated sequences were grouped. (B) Model for the evolution of gnathostome Dacts. The pairwise grouping of Dact1/3 and Dact2/4 and their associated genes suggests that after the first vertebrate genome duplication (1R), a Dact1/3 and a Dact2/4 precursor was generated, which during the 2R gave rise to the individual Dact1, Dact3, Dact2 and Dact4 genes. Subsequently, in Dact3 the leucine zipper required for Dact dimerization was reduced. Moreover, in teleosts, motif 4b located in the center of the Tcf3 interacting region was reduced (dact3b) or eliminated (dact3a). In Dact4, the leucine zipper as well as the PDZ binding domain of motif 11 g was lost, motifs 11d-f (Vangl binding domain) were reduced, and motifs 6 and 10 were gained. This suggests that Dact3 and, more prominently, Dact4 proteins have altered molecular properties compared to Dact1, Dact2, and the original dact.
