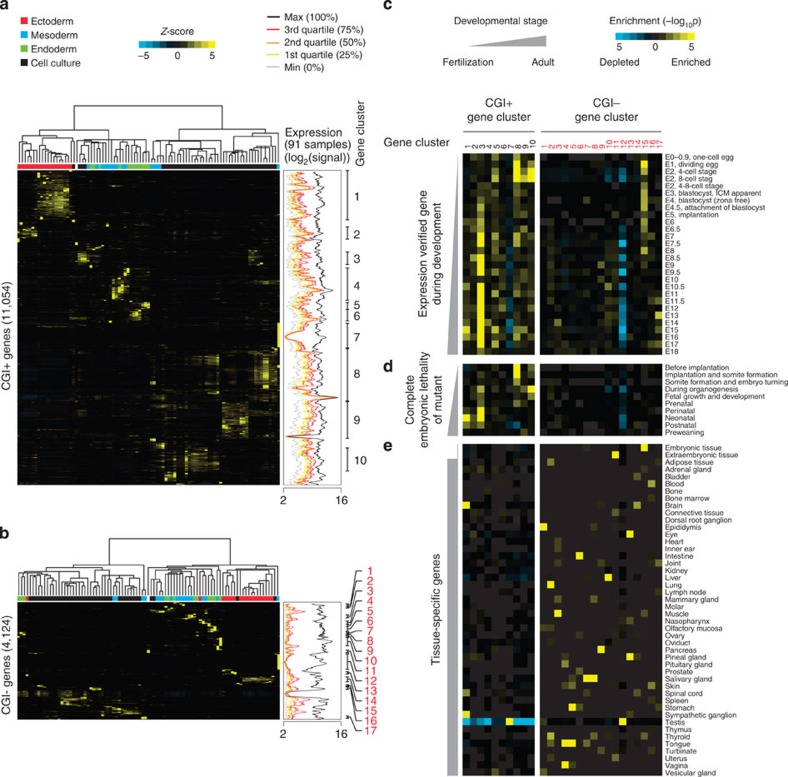
Figure 4. Functional segregation between CGI+ and CGI− genes during early embryo development.
(a,b) Unsupervised hierarchical clustering of gene expression variances (Z-scores) for CGI+ (a) and CGI− genes (b) from 91 tissues or cell lines (http://biogps.gnf.org34; left clustering result). For each sample, the putative origin of embryonic germ layer is indicated with a different colour (upper part of heatmap). Moving average plots in the middle (window size: 64, bin size: 1; genes are ordered as in gene cluster results) show global gene expressions of 91 different tissues or cell lines as shown in Fig. 2c. Gene clusters (gene sets) showing a similar pattern of expression variances are defined, and indicated as numbers in black and red (CGI+ and CGI− genes, respectively; right end). (c–e) Heatmaps showing hypergeometric P values calculated by gene set overlap analyses between gene sets from hierarchical clustering (Fig. 4a,b) and expression verified gene sets during embryo development (c), gene sets showing complete embryonic lethality upon deletion (d) or tissue-specific genes (e) (see Methods section for detail).