Figure 2.
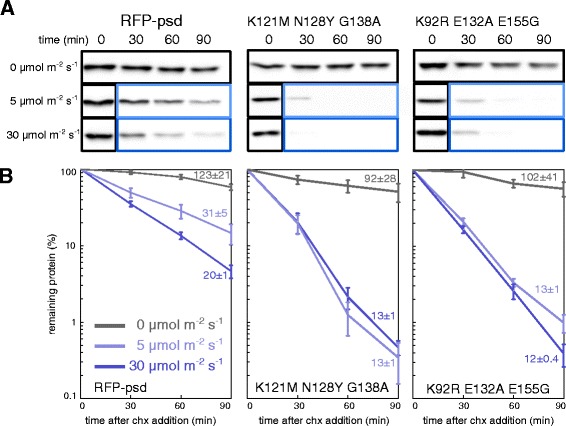
Quantification of psd module variant behavior. A) Yeast cells expressing PADH1 -RFP-psd (plasmid based) or the variants K121M N128Y G138A and K92R E132A E155G were grown in liquid medium in darkness. After removal of the first sample (t=0 hours), cycloheximide (chx) was added to stop protein synthesis; cells were kept in the dark (black box) or exposed to blue light (LED lamp, 465 nm, light blue box: 5 μmol m-2 s-1, dark blue box: 30 μmol m-2 s-1) for the rest of the experiment. Equal amounts of sample were collected at the indicated time points and subjected to alkaline lysis and western blotting. B) Quantification of the immunoblots shown in A. Curves are the means obtained from at least four independent measurements (error bars: s.e.m.). The half-lives (±standard error) that are indicated next to each curve were obtained by fitting the data to an exponential decay using the software Origin 7.
