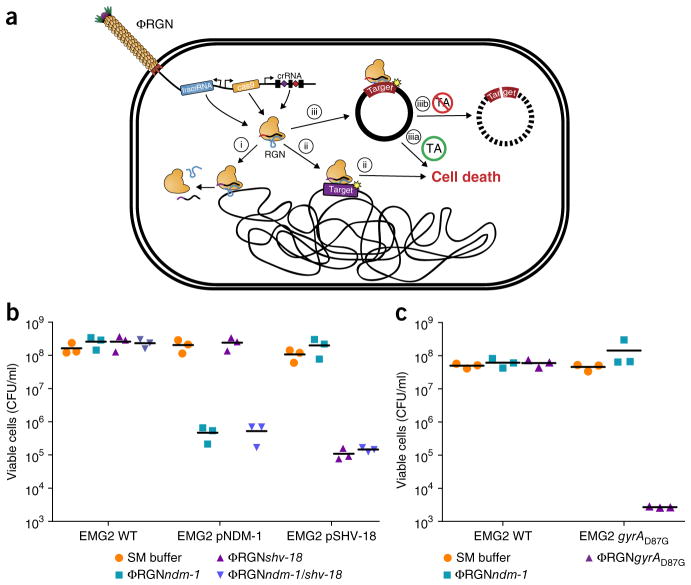
Figure 1.
RGN constructs delivered by bacteriophage particles (ΦRGN) exhibit efficient and specific antimicrobial effects against strains harboring plasmid or chromosomal target sequences. (a) Bacteriophage-delivered RGN constructs differentially affect host cell physiology in a sequence-dependent manner. If the target sequence is: (i) absent, the RGN exerts no effect; (ii) chromosomal, RGN activity is cytotoxic; (iii) episomal, the RGN leads to either (iiia) cell death or (iiib) plasmid loss, depending on the presence or absence of toxin-antitoxin (TA) systems, respectively. (b) Treatment of EMG2 wild-type (WT) or EMG2 containing native resistance plasmids, pNDM-1 (encoding blaNDM-1) or pSHV-18 (encoding blaSHV-18), with SM buffer, ΦRGNndm-1, ΦRGNshv-18, or multiplexed ΦRGNndm-1/shv-18 at a multiplicity of infection (MOI) ~20 showed sequence-dependent cytotoxicity as evidenced by a strain-specific reduction in viable cell counts (n = 3). CFU, colony-forming units. (c) E. coli EMG2 WT or EMG2 gyrAD87G populations were treated with SM buffer, ΦRGNndm-1 or ΦRGNgyrAD87G at MOI ~20, and viable cells were determined by plating onto Luria-Bertani agar (n = 3).