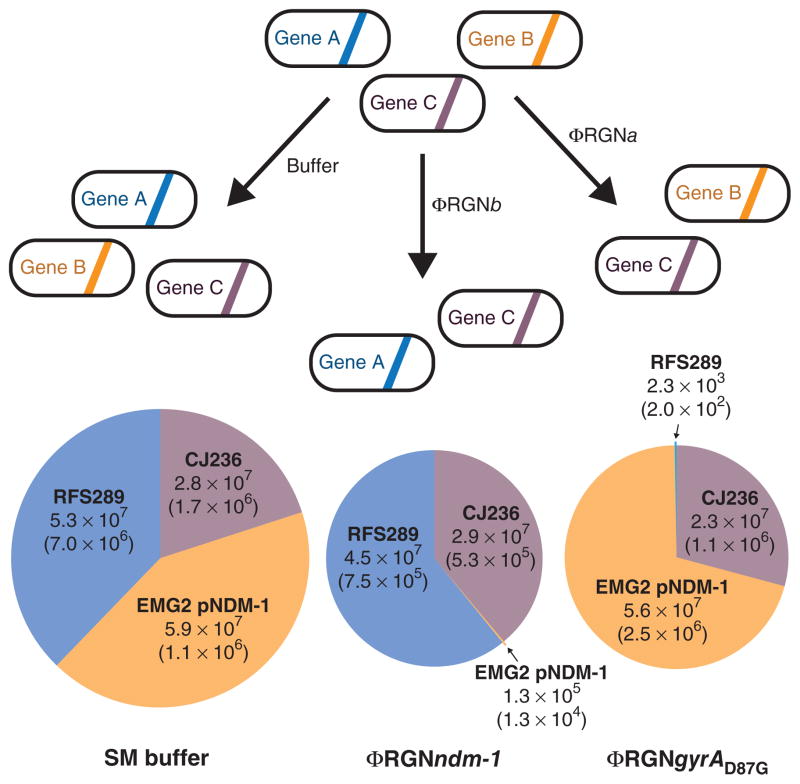
Figure 4.
Programmable remodeling of a synthetic microbial consortium. A synthetic population composed of three different E. coli strains was treated with either SM buffer, ΦRGNndm-1, or ΦRGNgyrAD87G at an MOI ~100 and plated onto LB with chloramphenicol, streptomycin or ofloxacin to enumerate viable cells of E. coli CJ236, EMG2 pNDM-1 or RFS289 strains, respectively. ΦRGNndm-1 targets blaNDM-1 in EMG2 pNDM-1 and ΦRGNgyrAD87G targets the gyrAD87G allele in RFS289. Circle area is proportional to total population size and numbers represent viable cell concentrations (CFU/ml) of each strain after the indicated treatment. The s.e.m. based on three independent experiments is indicated in parentheses (n = 3).