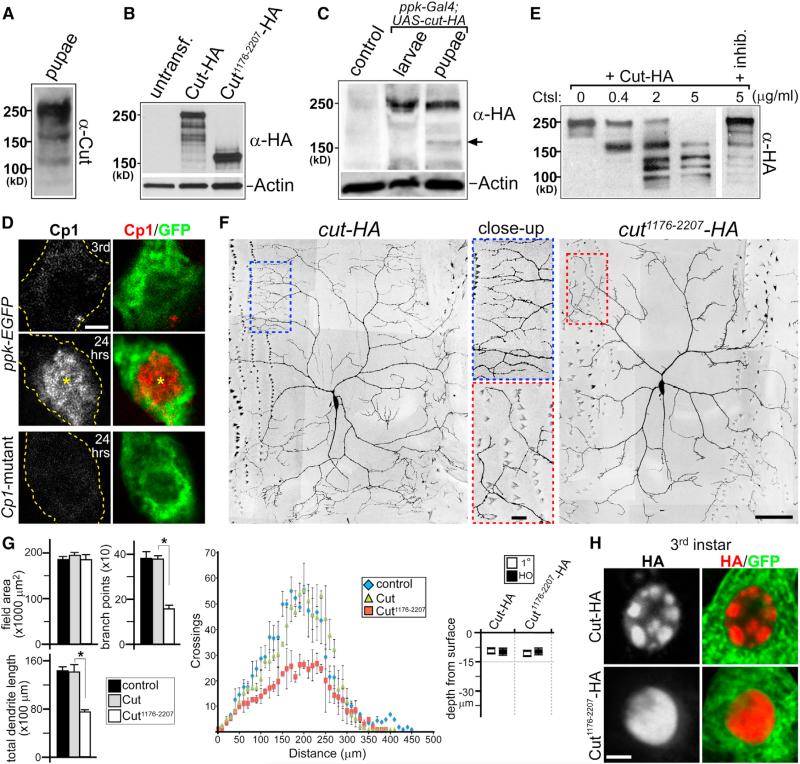
Figure 4. Context-Dependent Production and Function of Truncated Cut Isoform.
(A–C) Western blot analyses of Cut protein isoforms.
(A) Detection of Cut proteins from whole-animal lysate at 24 hr APF.
(B) Cut-HA and Cut1176-2207-HA protein expression in S2 cells. Note that the major protein bands in each lane, corresponding to intact Cut-HA and Cut1176-2207-HA, are near 250 and 160 kDa in size, respectively. untransf., untransfected.
(C) Cut-HA expression in ppk-Gal4; UAS-cut-HA third-instar larvae and 24 hr APF pupae lysates. Note the appearance of near 160 kDa protein isoform in pupal stage (arrow). Lysate from 24 hr APF UAS-cut-HA pupae (no ppk-Gal4) is the control.
(D) Cp1 plus GFP IHC antibody staining of wild-type ddaC neurons (ppk-EGFP) in third-instar larvae (Third), 24 hr APF pupae (24 hr), and Cp1-mutant ddaC neuron at 24 hr APF. Each image represents a single confocal plane. Note the nuclear Cp1 staining at 24 hr APF (*) in wild-type ddaC neuron.
(E) In vitro cleavage assay using purified Cut-HA protein and Ctsl in increased protease concentrations. Cleavage reaction is sensitive to Ctsl inhibitor Z-FF-FMK (inhib.) (50 μM).
(F) Fixed-tissue images of ddaC neurons from third-instar larvae expressing either Cut-HA or Cut1176-2207-HA. Color-dashed boxes are shown as corresponding close-up panels.
(G) Quantitative analyses of third-instar ddaC neuron dendrite phenotypes: field area, branchpoints, dendrite length, Sholl analysis, and depth from surface (n = 6 in all groups). *p < 0.005, Wilcoxon two-sample test. Error bars represent SEM.
(H) Cut-HA and Cut1176-2207-HA nuclear staining patterns in third-instar larvae ddaC neurons (ppk-Gal4; UAS-mCD8::GFP; UAS-cut-HA or UAS-cut1176-2207-HA).
Scale bars, 2 μm (D and H) and 50 μm (F). See also Figure S4.