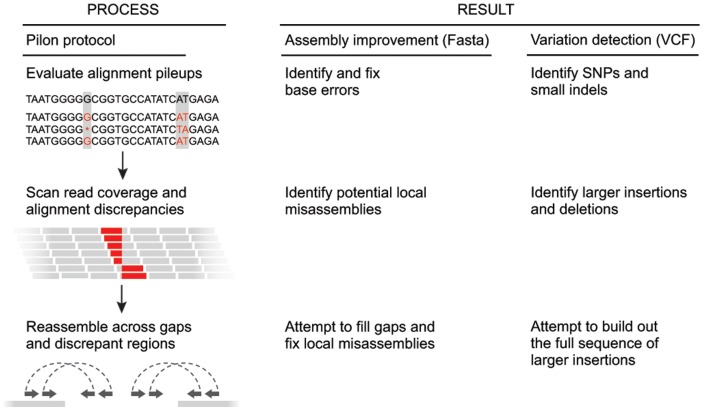
Figure 1. Simplified overview of the Pilon workflow for assembly improvement and variant detection.
The left column depicts the conceptual steps of the Pilon process, and the center and right columns describe what Pilon does at each step while in assembly improvement and variant detection modes, respectively. During the first step (top row), Pilon scans the read alignments for evidence where the sequencing data disagree with the input genome and makes corrections to small errors and detects small variants. During the second step (second row), Pilon looks for coverage and alignment discrepancies to identify potential mis-assemblies and larger variants. Finally (bottom row), Pilon uses reads and mate pairs which are anchored to the flanks of discrepant regions and gaps in the input genome to reassemble the area, attempting to fill in the true sequence including large insertions. The resulting output is an improved assembly and/or a VCF file of variants.