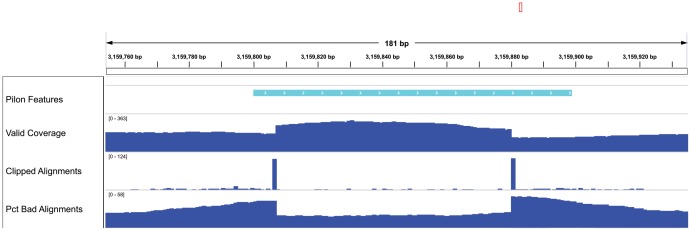
Figure 2. Example Pilon generated genome browser tracks.
This region was flagged by Pilon as containing a possible local mis-assembly, but Pilon was unable to determine a fix due to a tandem repeat sequence. The tracks shown here include: Pilon Features track indicating the extent of the region flagged by Pilon as containing a potential mis-assembly, Valid Coverage track indicating the sequence coverage of valid read pair alignments excluding the clipped portions of the alignments, Clipped Alignments track indicating the number of reads soft-clipped at each location, Pct Bad Alignments track indicating the percentage of the total reads aligned to each location which are not part of Valid Coverage. These tracks are created with the ‘—tracks' command-line option. Together, these tracks reveal the true bounds of the mis-assembly, and indicate that there are likely missing copies of the tandem repeat in the draft assembly. In this case, manual analysis revealed the draft assembly was missing two of three full copies of a 57-base tandem repeat.