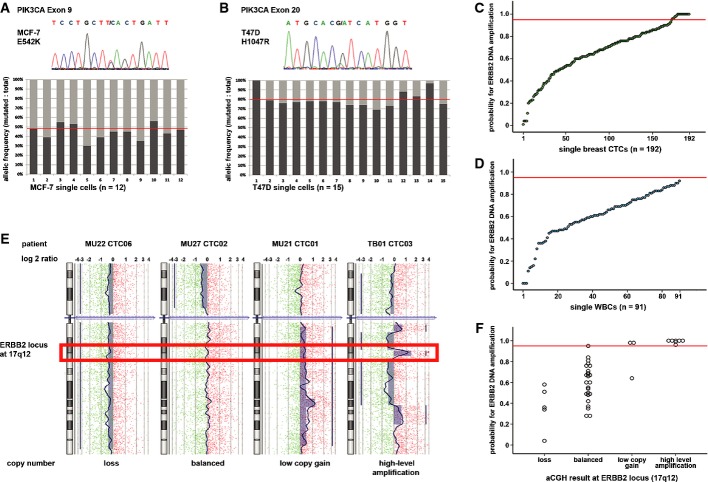
Figure 3. Assays for molecular single cell analysis.
- Exon 9 mutation E545K was detected in all single MCF-7 cells. The mutant allele is representing 45% of detected sequences averaged over all analyzed single cells and 48% in genomic DNA of MCF7 (right). The horizontal red line indicates the allelic ratio of non-amplified genomic DNA.
- Exon 20 mutation H1047R was detected in all single T47D cells. The mutant allele is representing 80% of detected sequences averaged over all analyzed single cells, as well as in genomic DNA. Cell 01 shows an allelic loss of the wild-type sequence. The horizontal red line indicates the allelic ratio of non-amplified genomic DNA.
- ERBB2 copy numbers were assessed by qPCR in 192 CTCs from breast cancer patients. Twenty-one single cells of 7 of 42 patients displayed an amplification probability above 95% (indicated by the red horizontal line).
- ERBB2 amplification qPCR determined all single WBCs (n = 91) to be negative for ERBB2 amplification (below the red horizontal line).
- High-resolution aCGH profiles of four individual cells showing DNA loss (left), balanced aCGH profile (second from left), low copy number gain (second from right) and high-level amplification (right) at ERBB2 locus (hybridization ratio for single probes shown on a log2 scale).
- ERBB2 copy number by aCGH correlates with amplification probability score by qPCR. A qPCR amplification probability score ≥ 0.95 (red horizontal line) indicates ERBB2 amplification. Two samples dropped out of analysis due to failed amplification of qPCR fragments.