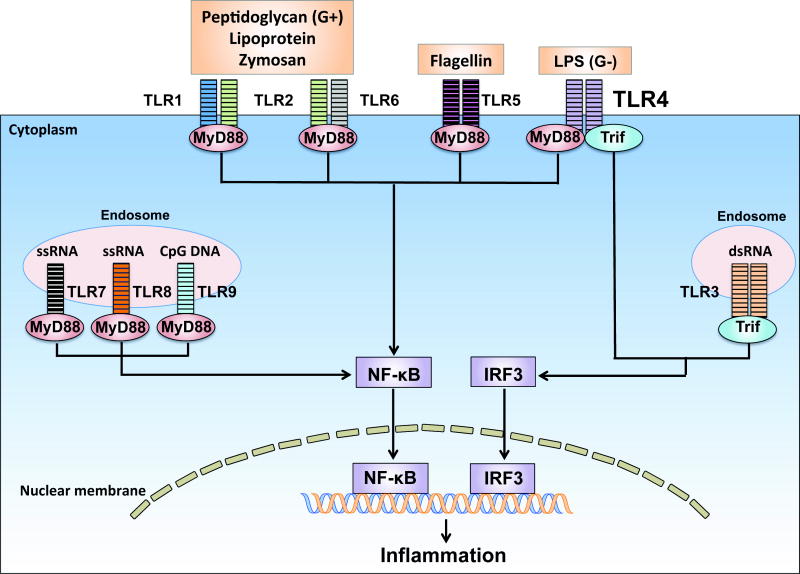
Fig. 1. Pathogen sensing by Toll-like receptors.
Toll-like receptors (TLRs) are pattern-recognition receptors. All TLRs are transmembrane proteins. Some TLRs such as TLR1, 2, 4, 5 and 6 are expressed on the cell surface whereas others such as TLR3, 7, 8 and 9 are located almost exclusively in intra-cellular compartments such as endosomes. Different TLRs recognize different microbial components 49. For example, TLR4 senses lipopolysaccharide (LPS), a wall component of Gram-negative (G−) bacteria such as Escherichia coli. TLR2 recognizes lipoprotein, a wall component of Gram-positive (G+) bacteria such as Staphylococcus aureus or Streptococcus pneumoniate. TLR2 heterodimerizes either with TLR1 to recognize triacylated lipopeptide or with TLR6 to recognize diacylated lipopeptides. TLR5 senses bacterial flagellin, a protein component of flagella. TLR3 recognizes viral double-stranded RNA (dsRNA), whereas TLR7 and 8 are the sensors for single stranded RNA (ssRNA). Finally, TLR9 senses bacterial CpG-rich hypomethylated DNA (CpG DNA) motifs. With the exception of TLR3, all TLR members signal through the adaptor myeloid differentiation primary-response gene 88 (MyD88) to recruit the downstream kinases. TLR3 signals through the adaptor TIR-domain-containing adaptor protein inducing interferon-β (IFN-β)-mediated transcription-factor (Trif). TLR4 signals through both MyD88 and Trif dependent pathways. Activation of these signaling pathways ultimately activates the transcription factors such as nuclear factor-κB (NF-κB) and IFN regulatory factor 3 (IRF3), which leads to production of diverse proinflammatory cytokines 50.