Figure 3.
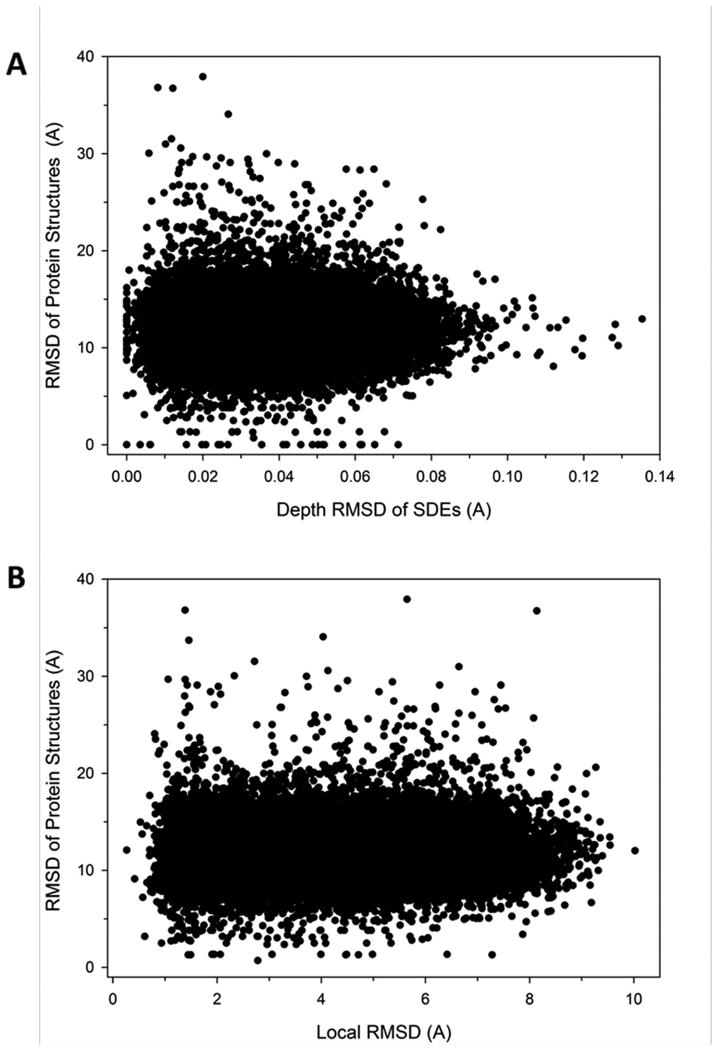
Global structural similarity of proteins that have similar side-chain environments. (A) Global coordinate RMSD of protein structures whose residue has similar SDEs. For each residue in the reference protein database, five most similar residues to the query residue in terms of SDEs were selected. Their depth RMSD of the side-chains in a sphere of 8.0 Å and the global RMSD of the whole protein structures were computed. Global RMSD was computed with BioShell, which computes RMSD of gapped structure alignment between two proteins. (B) For the same residue pairs shown in the plot A, the conventional RMSD of the neighboring side-chain centroids were compared with the global RMSD of the proteins.
