Figure 4.
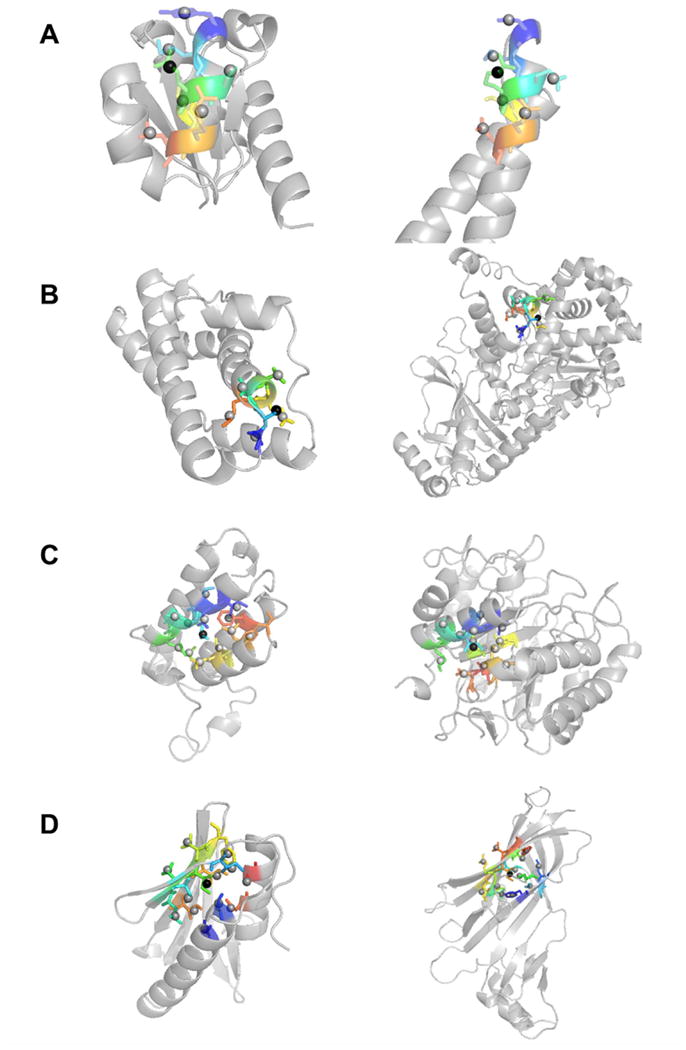
Examples of similar SDEs. Residues included in SDEs are shown in color. (A) SDE of residue 64 (Glu) of IIBcellobiose (PDB code: 1iib) (left) and residue 146 (Arg) of RNA polymerase II mediator complex protein MED7 (1ykh, chain A) (right). The center side-chain is shown in black spheres. 8.0 Å was used for the sphere to define SDEs. Both SDEs contain seven side-chain centroid points of neighboring residues (shown in the stick representation). The residues are 60, 61, 63, 64, 65, 67, and 68 for 1iib and 32, 33, 35, 36, 37, 39, and 40 from 1ykhA. The depth RMSD (dRMSD) of the two SDEs is 0.022 Å. (B) Residue 40 (Ser) in fertilization protein (1lis) and residue 312 (Ser) of Anthrax lethal factor (1yqyA). Six residues are included: residue 39, 40, 41, 42, 43, 44 for 1lis and 311, 312, 313, 314, 315, 316 from 1yqyA dRMSD: 0.023 Å. (C) SDEs of residue 12 (Leu) in bromodomain of GCN5 (PDB: 1e6i) and residue 71 (Ile) of β-mannose (1bqcA). 13 residues are in the SDEs, 8, 9, 11, 12, 13, 15, 16, 46, 49, 50, 53, 58, and 64 for 1e6i and 67, 68, 70, 71, 72, 74, 75, 81, 83, 113, 114, 117, and 121 for 1bqcA The depth RMSD is 0.037 Å. (D) Residue 41 (Ile) of map kinase 14 (1ew4) and residue 196 (Ile) of tetracycline repressor (2bj0A). 13 residues are included. 1ew4: 39, 40, 41, 42, 43, 17, 21, 30, 32, 49, 51, 91, 95; and 2bj0A 194, 195, 196, 197, 198, 31, 33, 50, 137, 139, 161, 175, 177. dRMSD: 0.035 Å.
