FIGURE 1.
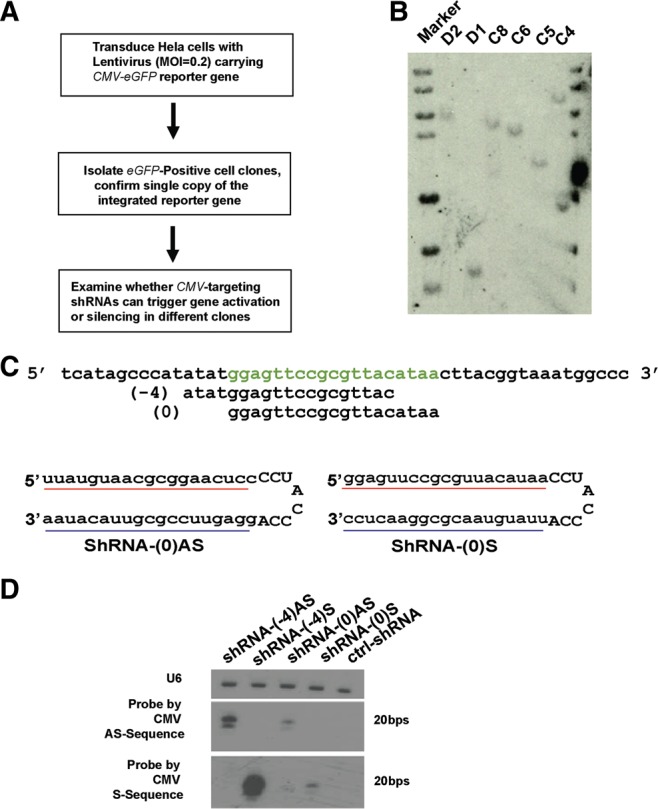
A cell model to evaluate the effect of local chromatin environment on shRNA-mediated TGA. (A) Experimental design for analyzing TGA in HeLa clones. (B) Southern blot to analyze if isolated cell clones, such as D2, D1, C8, C6, C5, and C4, have one copy of the CMV-EGFP reporter gene integrated into the genome. All clones were isolated from the same transduced HeLa cell population. (C) The design of shRNAs targeting the CMV promoter. The sequence in green is denoted as site 0, which corresponds to the region −531 to −513 bp upstream of the transcriptional starting site (TSS). The numbers in parentheses denote the CMV promoter region targeted by each shRNA. The positions of the bottom (guide) and upper (passenger) strands in shRNA-(0)S (lower right) are reversed in shRNA(0)AS (lower left). (D) Northern blot to analyze preferential selection of the bottom shRNA strands as the guide strands. HeLa cells were transfected by the plasmids expressing these shRNAs (see Materials and Methods for details). An irrelevant shRNA is used as the negative control (ctrl-shRNA). U6 RNA was probed for the loading control. The images were taken from the same membrane hybridized with different probes.
