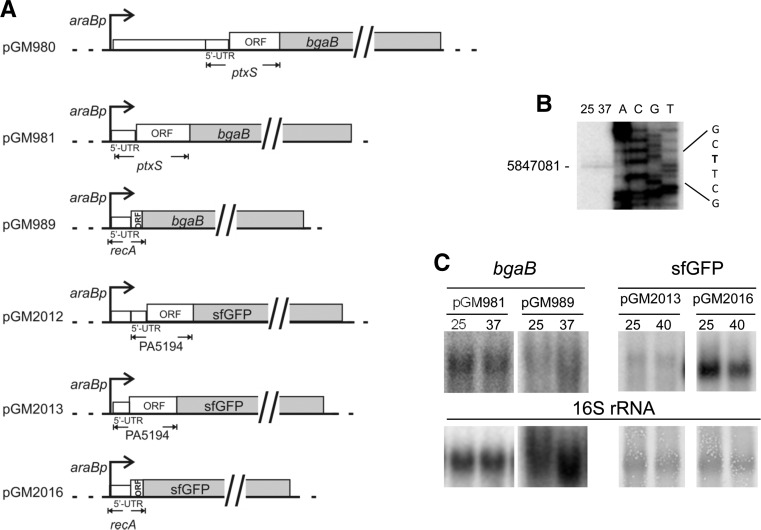
FIGURE 3.
Constructs used for validation of putative RNATs and transcriptional analysis. (A) Map of plasmids encoding PtxS-BgaB and PA5194-sfGFP. Control plasmids carrying the 5′ end of the E. coli recA gene in frame with the reporter genes are also represented. Details about plasmid construction and coordinates of the cloned regions are reported in Materials and Methods. Pseudomonas aeruginosa and E. coli 5′ UTRs and ORFs are drawn to scale. (Dotted lines) vector sequences; (bent arrows) araBp promoter; (empty boxes) P. aeruginosa and E. coli DNA; (gray boxes) reporter genes. (B) Mapping the 5′ end of the PA5194 mRNA by primer extension. Primer extension was performed with the radiolabeled oligo 3003 on 30 µg of RNA extracted from exponential cultures of PAO1 grown at 25°C and 37°C. The same oligonucleotide was used for Sanger-sequencing of construct 3. The coordinate of the identified 5′ end and the sequence context (in bold the 5′ end T) are reported beside the panel. (C) Northern blot analysis of ptxS-bgaB and PA5194-sfGFP fusions in P. aeruginosa. Cultures of the PA01 strain carrying the plasmids indicated above the lanes were grown at 25°C up to OD600 = 0.5. The cultures were induced with 0.1% arabinose, split and further incubated at the temperature indicated above the lanes for 45 min. Northern blotting was performed as described in Materials and Methods after electrophoresis in a 1.5% denaturing agarose gel. The filter was hybridized with oligonucleotides specific for bgaB or sfGFP, as indicated on top of the panels. To check gel loading, the filters were either hybridized with the 16S rRNA-specific 1396 oligonucleotide (bgaB panels) or stained with methylene blue before the hybridization (sfGFP panels; the bands corresponding to 16S rRNA are shown). The transcript size was roughly estimated by comparison with rRNA migration as follows: pGM981 and pGM989, 2.5 kb; pGM2013, 1.2 kb; pGM2016, 1.1 kb.