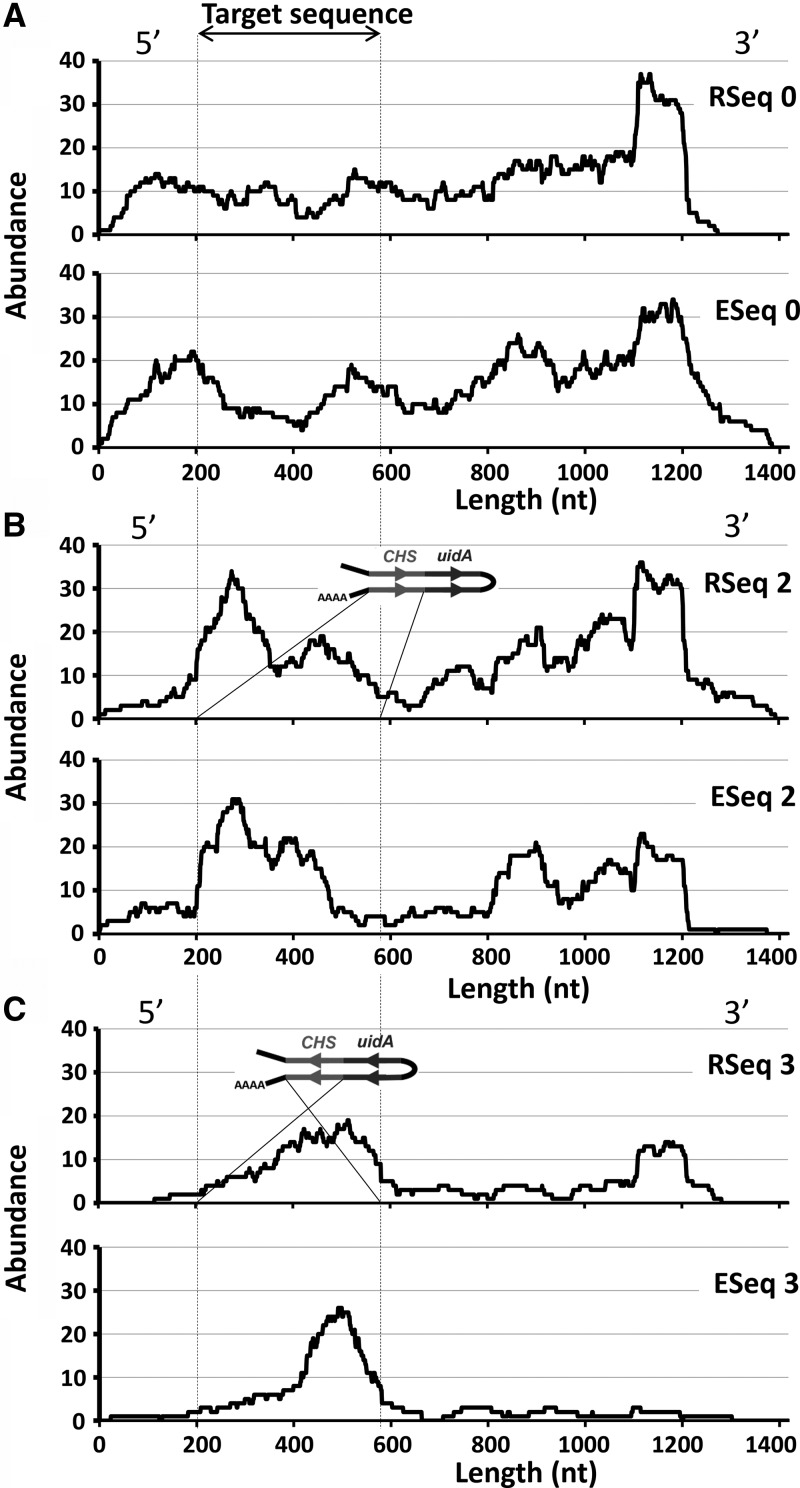
FIGURE 4.
Alignment of reads from RSeq and ESeq libraries to the sequence of AtCHS mRNA. (A) In wild-type plants the reads were distributed across the entire length of AtCHS mRNA sequence; treatment with 5′PDE (ESeq 0) resulted in a slight increase in the coverage. (B) In RSeq 2, made from the plants transformed with chsS-gusS^gusA-chsA and least efficient in silencing of AtCHS mRNA, the read coverage upstream (5′) of the target sequence is clearly reduced, but the coverage downstream (3′) is comparable to that in wild-type plants. The reads corresponding to the 3′ fragment originated from RNA sensitive to 5′PDE treatment, as indicated by reduced coverage in ESeq 2. (C) Even greater reduction in AtCHS mRNA-derived reads was observed among plants transformed with chsA-gusA^gusS-chsS. In both (B) and (C) the transgene-derived reads are more abundant toward the predicted poly-A tail within the ihpRNA structure. Coverage indicates number of corresponding reads per nucleotide out of 9 million total reads per library.