Abstract
Background
Epigenetic alterations are well documented in hepatocarcinogenesis. However, hypomethylation of long interspersed nuclear element 1(LINE-1) promoter and its relationship with clinicopathological features in hepatocellular carcinoma (HCC) remain unknown.
Methods
The bisulfite-specific PCR and DNA sequencing analysis was performed to assess the methylation status of LINE-1 promoter in a pilot cohort of 71 patients with HCC. Additionally, methylation levels of two hot CpG sites of LINE-1 promoter, site 7 and 18 were measured by real-time PCR and compared with clinicopathological parameters in a cohort of 172 HCC. All the patients included were in BCLC stage A or B.
Results
Most patients with HCC (87.3%) showed hypomethylation of LINE-1 promoter compared with HBV-related cirrhosis and normal controls (P < 0.001). The HCC patients with LINE-1 promoter hypomethylation had a median tumour-free survival (TFS) and overall survival (OS) post-resection of 22.0 (95% CI: 13.3–30.7) months and 35.0 (95% CI: 24.0–46.1) months, respectively, compared with 40 months and ∼60 months for those with LINE-1 promoter hypermethylation (P < 0.05). Multivariate analyses showed that the hypomethylation level at CpG site 7 and 18 of LINE-1 promoter, along with tumour size and tumour differentiation, was independently associated with both TFS and OS for patients with HCC after resection.
Conclusion
Promoter hypomethylation of LINE-1, especially at the CpG site 7 and 18, was associated with a poor prognosis in HCC.
Keywords: Hepatocellular carcinoma, long interspersed nuclear element-1, methylation
Hepatocellular carcinoma (HCC) is one of the most common cancers in the Asian and Pacific regions 1. Owing to its aggressiveness and high rate of post-operative recurrence, the overall prognosis of patients with HCC remains unsatisfactory after hepatectomy and loco-regional ablation 2,3. Although many studies have been published in an effort to explore suitable biomarkers for the prediction of the prognosis and the recurrent rates after resection of HCC, clinical features including stage and grade still play an important role in determining treatment and prediction of recurrence 4. However, there is significant variability in the prognosis of patients with similar clinical characteristics. A reliable prognostic marker that accurately predicts the outcome of HCC after hepatectomy will help select the optimal treatment modalities for individual patients and a strategy for monitoring the disease.
Genetic and the epigenetic alterations accumulate during the initiation, promotion and progression of HCC 5. These changes included mutations of oncogenes and tumour suppressor genes, DNA rearrangements associated with HBV, insertion or deletion of chromosome regions and loss of heterozygosity and hypermethylation of promoter regions in the tumour suppressor genes 6. Interestingly, a global hypomethylation has recently been postulated to be an important contributor to HCC tumourigenesis.
The long interspersed nuclear element-1 (LINE-1,Fig.1A), 6 kd in size, is one of the most important parts of repetitive DNA elements and constitutes about 17% of the human genome sequence 7,8, but most LINE-1s are inactive because of 5′truncations, rearrangement and mutations 7. There are only ∼100 copies of retrotransposition competent full-length LINE-1 containing promoter region and two ORFs, and the methylation of LINE-1′s promoter can regulate the expression of LINE-1 9. While the methylation of LINE-1 promoter region limits its expression, hypomethylation might lead to an increase in expression and retrotransposition of this gene 10–12. Retrotransposition of LINE-1 gene may lead to chromosomal instability, DNA rearrangement and the alteration of ectopic gene expression in cancerous tissues13,14. Similar to a previous report 15, our recent study demonstrated that LINE-1 could promote human cancer cell lines growth with an invasive potential 16. It has also been reported that LINE-1 hypomethylation could be used as a new marker of the HCC for diagnoses and to guide treatment strategies 17,18, especially the hypomethylation of LINE-1′s promoter is found to be closely associated with the progress of tumour 19. Previous studies focused mostly on the genome-wide hypomethylation; however, the association of hypo-methylation of the LINE-1 promoter with the prognosis of patients with HCC remains unknown. To address these questions and further evaluate the role of LINE-1 activation by hypomethylation in the tumourigenesis of HCC, we evaluated the LINE-1 promoter methylation status at each CpG site in a relatively large clinical cohort of HCC patients with HCC and chronic HBV infection. We also assessed the associations between the levels of LINE-1 promoter hypomethylation and clinicopathological parameters including tumour size and differentiation as well as the patient outcomes after curative resection in Chinese patients with HCC.
Figure 1.
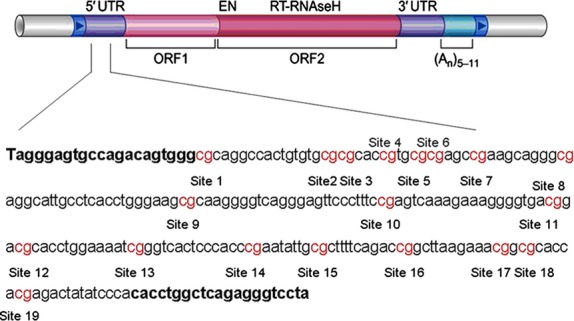
Schematic diagram of LINE-1. LINE-1 DNA fragments comprise a 5′-untranslated region (5′-UTR) with internal promoter activity. The sequence shown represents a 247-base pair fragment (base pairs 117–364) in the promoter of LINE-1. Numbers 1–19 refer to locations of the CpG site within the LINE-1 elements tested.
Materials and methods
HCC Patients selection, clinical characteristics and tissue sampling
A total of 301 patients with HBV-related HCC were included in this study. Patients were identified prospectively and consecutively. HCC and non-tumourous tissues were obtained from surgical resections from December 2005 to December 2008. Diagnosis of HCC was based on the criteria from European Association for the Study of the Liver 20. According to the Barcelona Clinic Liver Cancer (BCLC) staging classification system 4, there were 153 patients in stage A and 148 in stage B. A total of 36 patients had an incomplete resection with microscopic tumour evidence at the surgical margin, 12 died from unrelated causes without recurrence of HCC and 10 were lost to follow-up for non-medical reasons and were excluded from this study. Therefore, 243 patients with HCC were included in the final analysis (Table1). In addition, liver tissue from 29 patients with chronic HBV-related cirrhosis (child-pugh A/B: 26/3), 9 hepatic hemangiomas and 10 benign hepatic cysts without viral hepatitis or cirrhosis were included as control. The selected tissues were snap-frozen in liquid nitrogen and stored at −70°C until DNA extraction. The clinicopathologic parameters were examined prospectively without the knowledge of molecular genetic results.
Table 1.
The clinicopathological characteristics of 243 patients with HCC
| Variable | Cohort 1 (N = 71) (%) | Cohort 2 (N = 172) (%) | ||||
|---|---|---|---|---|---|---|
| N | % | N | % | |||
| Age (Mean age ± SD)(y),range | 49.08 ± 7.73 (18–62) | 46.28 ± 8.87 (28–71) | ||||
| Gender | ||||||
| Male | 54 | 76.06 | 150 | 87.21 | ||
| Female | 17 | 23.94 | 22 | 12.79 | ||
| Viral infection | ||||||
| HBV DNA-positive | 46 | 64.79 | 105 | 61.05 | ||
| HBeAg-positive | 28 | 39.44 | 66 | 38.37 | ||
| Child-Pugh classification | ||||||
| Class A | 62 | 87.32 | 161 | 93.60 | ||
| Class B | 9 | 12.68 | 11 | 6.40 | ||
| Tumour size | ||||||
| <3.8 cm | 33 | 46.47 | 93 | 54.07 | ||
| ≥3.8 cm | 38 | 53.52 | 79 | 45.93 | ||
| Number of nodules | ||||||
| Single | 63 | 88.73 | 144 | 83.72 | ||
| ≥2 | 8 | 11.26 | 28 | 16.28 | ||
| Tumour differentiation | ||||||
| well | 10 | 14.08 | 35 | 20.35 | ||
| mediately | 18 | 25.35 | 58 | 33.72 | ||
| poorly | 43 | 60.56 | 79 | 45.93 | ||
| BCLC stage | ||||||
| stage A | 36 | 50.7 | 81 | 47.09 | ||
| stage B | 35 | 49.29 | 91 | 52.91 | ||
| AFP | ||||||
| ≤400 ng/ml | 48 | 67.61 | 96 | 55.81 | ||
| >400 ng/ml | 23 | 32.39 | 76 | 44.19 | ||
The study protocol was approved by the 302nd Hospital Research Ethics Committee and a written informed consent was obtained from all participants. None of the patients had received prior treatment for HCC, including radiation or chemotherapy. Patients were followed up every 2 months during the first post-operative year and at approximately three to four monthly intervals thereafter until June 2011. Routine evaluation of patients included physical examination, chest roentgenography, blood chemistry analysis, HBV DNA levels and measurement of tumour markers (carcinoembryonic antigen and alpha-fetoprotein). Chest and abdominal computed tomography, brain magnetic resonance imaging and bone scintiscan were performed every 6 months for 3 years after surgery. Additional investigations were performed whenever any symptoms or signs of recurrence developed.
DNA extraction, bisulfite modification in DNA and Long interspersed nuclear element-1 (LINE-1) methylation analyses
All samples were detected for LINE-1 methylation analyses without knowledge of the clinicopathological or follow-up data. Genomic DNA was extracted with the Blood and Tissue DNA Kit (QIAGEN, Dusseldorf, Germany). A total of 200 ng of genomic DNA from each sample was treated with sodium bisulfite 21. Briefly, after denaturation in 0.3 M NaOH at 37°C for 15 min, sodium bisulfite (3.1 M) and hydroquinone (0.5 mM) were added. The reaction was performed at 50°C for 16 h followed by desalting was then conducted by using Wizard DNA purification resin (Promega, Madison, WI, USA). After Bisulfite modification, DNA samples were treated with 0.3 M NaOH at 37°C for 15 min. Modified DNA was precipitated with ethanol, washed in 70% ethanol, dried and resuspended in 20 μl of distilled water.
LINE-1 bisulfite sequencing analysis
To obtain the overall DNA methylation status of the LINE-1 promoter region, patients were randomly divided into two cohorts: cohort 1 (71 HCCs) and cohort 2 (172 HCCs). DNA methylation status was initially measured in cohort 1 along with 19 normal liver tissues and 29 cirrhosis tissues by bisulfite seq-uencing analysis. We used the following primers: F-5′-TAGGGAGTGTTAGATAGTGA-3′ and R- 5′-TAAAACCCTCTAAACCAAATA-3′ to amplify the fragment from 117 to 364 bp of the LINE-1 promoter region (Gen Bank: X58075.1), which covers the entire 19 CpG sites (Fig.1). The 50 ng of bisulfite-treated genomic DNA was used in the PCR reaction. The 50 μl reactions for LINE-1 promoter were run for 30 cycles as follows: predenaturation at 95°C for 30 sec, denaturation at 95°C for 5 sec, annealing 50°C for 30 sec and extension at 72°C for 30 sec. Amplified bisulfite-sequencing LINE-1 promoter fragments were cloned into the pEASY-T1 vector (TransGen Biotech, Beijing, China). Twenty clones from each sample were randomly selected for DNA sequencing. Sequencing analysis was performed by Shanghai Invitrogen Biotech Co. Ltd (Shanghai, China). To obtain the actual methylation status of each CpG site, we used the percentage of methylation of each CpG site in a given sample as a parameter, which was calculated as the number of methylations at a specific CpG site divided by the total number of clones which were sequenced.
Quantitative methylation analysis of LINE-1
To further characterize the association between LINE-1 promoter methylation and clinicopathological parameters, cohort 2 (172 HCCs) was used to verify methylation status of the CpG site 7 and 18 within the LINE-1 promoter. The methylation level of CpG site7 and 18 was evaluated using quantitative real-time PCR. The primers were used as follows:
| Methylated primers | Unmethylated primers | |
|---|---|---|
| Site7 | F:5′- AGGAATAGTTTYGGTTTATAG -3′ | F:5′- AGGAATAGTTTYGGTTTATAG -3′ |
| R:5′-ACAATACCTCRCCCTACTTCG-3′ | R:5′-ACAATACCTCRCCCTACTTCA-3′ | |
| Site18 | F:5′-TAGGGAGTGTTAGATAGTGA-3′ | F:5′-TAGGGAGTGTTAGATAGTGA-3′ |
| R:5′-ATAAAATATAATCTCRTAATACG-3′ | R:5′-ATAAAATATAATCTCRTAATACA-3′ |
The 100 ng of bisulfite-treated genomic DNA was used in the PCR by SYBR green PCR master mix (Takara, Otsu, Japan) and run on the Applied Biosystems Prism 7000 (Foster, CA, USA) real-time PCR machine. The 50 μl reactions for CpG site 7 and site 18 were run for 30 cycles as follows: predenaturation at 95°C for 5 min, denaturation at 95°C for 10 sec, annealing at 60°C for CpG site 7 (at 54°C for CpG site 18) for 20 sec and extension at 72°C for 20 sec and melt curve analysis from 65 to 95°C in 1°C increments. For the generation of standard curves, PCR products were purified through polyacrylamide gel electrophoresis. Resolved DNA bands were excised and eluted in 200 μl of pure water. Three-fold serial dilutions covering a 3–4 log dynamic range of eluted PCR products were used as templates in real-time qPCR to generate standard curves. GAPDH acted as an internal control. Real-time PCR reactions for unmethylated and methylated LINE-1 sequences were performed simultaneously in one 96-well plate. The percentage of methylation in a sample at site 7 or 18 of LINE-1 promoter was calculated using the formula: 100 × methylated reaction/(unmethylated reaction + methylated reaction).
Immunohistochemistry
Twenty-four paraffin-embedded HCC and surrounding non-tumourous tissues were used for immunohistochemical staining. Polyclonal rabbit anti-human antibody against LINE-1 (1:4000, provided by Professor Zhu YF) 22 was used. The interpretation of immune staining was performed by two independent pathologists without knowledge of clinical data. A positive score in a sample was measured as the percentage of positive cells in a total of 1000 carcinoma cells counted in five different fields (200 cells/field). The LINE-1 labelling index (LI) was calculated as the percentage of all tumour cells counted with nuclear staining. LI levels lower than 10% was described as negative.
Statistical analysis
Unsupervised hierarchical clustering analysis was carried out by using the ncss software. Statistical analyses were performed with sas 8.2 software (Cary, NC, USA). TFS and OS were compared using the log-rank test. Univariate and multivariate analyses were carried out based on the Cox proportional hazards regression model. The comparison among groups was performed using the X2-test and Student's t-test. A P value <0.05 was considered statistically significant.
Results
General findings
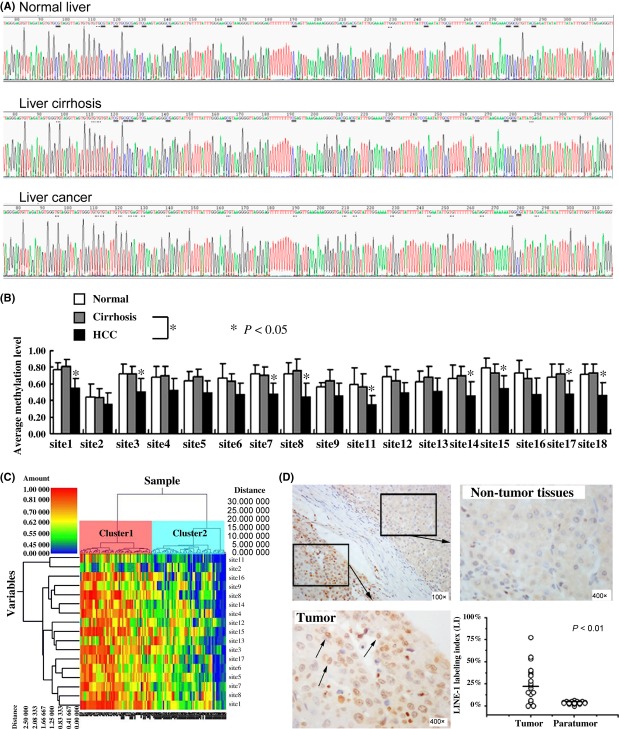
Because of a high rate of variation, the 10th and 19th CpG sites were excluded from this study. A total of 17 CpG sites in LINE-1 promoter were evaluated for methylation status. Methylation levels varied at different CpG sites (Fig.2), the lowest level was located at the 11th CpG (34%), whereas the first CpG sites at 5′ end had the highest methylation levels (55%). The methylation level in HCC at CpG sites 1, 3, 7, 8, 11, 14, 15, 17 and 18 was significantly lower than that of control liver tissues (P < 0.05). However, there was no significant difference in methylation status between the normal and cirrhosis samples at any CpG sites (P > 0.05). A hierarchical clustering analysis was performed according to the level of LINE-1 promoter methylation and the 119 cases (including 71 HCCs, 29 cirrhosis tissues and 19 normal liver tissues) were divided into two distinct subclasses, cluster 1 and cluster 2 (Fig.2C). The methylation level of cluster 1 was significantly higher compared with cluster 2. Among 57 cases of cluster 1 (57/119, 47.9%), there were 27 cases of cirrhosis (27/29, 93.1%) and 17 normal controls (17/19, 89.5%) and 13 HCCs (13/71, 18.3%). On the other hand, of the 62 cases of cluster 2 (62/119, 52.1%), there were 58 HCCs (58/71, 81.7%), two cases of cirrhosis (2/29, 6.9%) and two normal controls (2/19, 10.5%) (P < 0.001). These results strongly indicated a significant decrease in methylation levels of LINE-1 promoter in HCC compared with non-tumourous samples.
Figure 2.
Methylation Level of long interspersed nucleotide element-1(LINE-1) promoter and expression of LINE-1 in liver tissue. (A) Representative bisulfite sequencing analysis of the LINE-1 promoter in normal liver, cirrhosis and liver cancer. The methylated and unmethylated CpG sites corresponding to the 19 CpG sites are underlined in parallel, methylated CpGs were CG, whereas unmethylated CpGs were TG. (B) Methylation levels of all CpG sites in LINE-1 promoter were compared between samples from subjects with HCC or cirrhosis and normal liver control. *P < 0.05, relative to the respective control. (C) HeatMap showing unsupervised clustering of normal, cirrhosis and HCC samples (x-axis) between the methylation of all CpG (y-axis). Green corresponds to low methylation and Red to high methylation. (D) Representative samples of immunostaining for LINE-1 and quantitative analysis results for LINE-1-positive dots in HCC and corresponding non-tumourous liver tissues. LINE-1 staining in HCC showed nuclear positivity (arrows), but negative in non-tumourous liver tissues (× 400).
Expressions of LINE-1 by immunohistochemical staining
To demonstrate if hypomethylation of LINE-1 promoter leads to re-expression of LINE-1 in HCC, we explored the expression of LINE-1 in HCC and non-tumourous tissues by immunohistochemistry in 24 cases of HCC with hypomethylation of LINE-1 promoter. LINE-1 was expressed in HCC cells with positive nuclear staining and surrounding non-tumourous liver cells showed negative staining (Fig.2D).
Association of methylation level of LINE-1 promoter with outcomes after curative resection
To investigate the association between the level of LINE-1 promoter methylation status and outcomes after post-resection of HCC, the unsupervised clustering analysis was carried out in cohort 1 HCC. The 71 cases with HCCs were divided into two clusters by hierarchical clustering analysis. The methylation level of cluster 1 was higher than that of cluster 2 (Fig.3A). The median post-curative resection TFS was 40 months for patients in cluster 1, and their 1-, 3- and 5-year recurrence-free survival (RFS) rates were 80.6, 50 and 44.4% respectively. These were significantly longer than the median TFS of 22.0 (95% CI: 13.3–30.7) months in cluster 2, with their 1-, 3- and 5-year RFS rates of 65.71, 25.71 and 14.29% respectively (log- rank P = 0.0142, Fig.3B).
Figure 3.
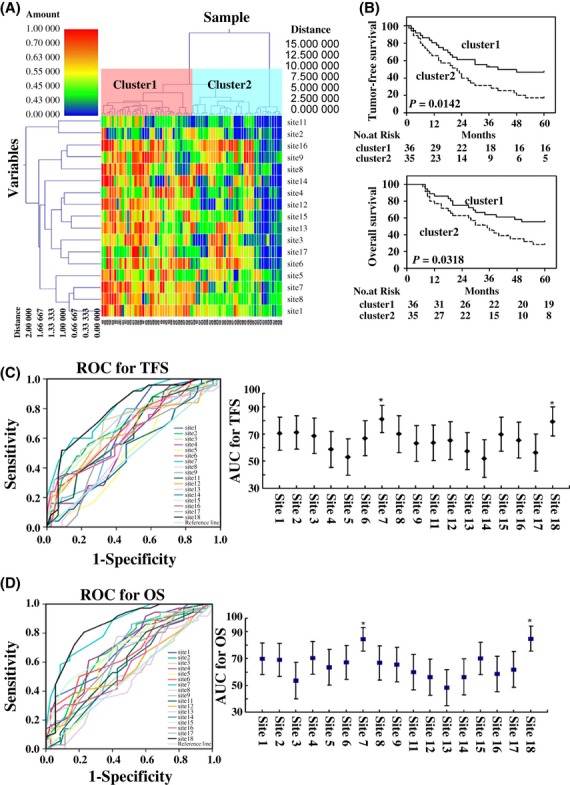
Unsupervised clustering based on LINE-1 promoter methylation level of HCC and outcomes of post-resection in cohort 1. (A) HeatMap showing unsupervised clustering of all HCC samples (x-axis) between the methylation of all CpG (y-axis). Green corresponds to low methylation and Red to high methylation. (B) TFS or OS curves of cluster 1 and cluster 2. TFS was significantly longer in cluster 1 than that of cluster 2 (P < 0.05). OS was significantly longer in cluster 1 than that of cluster 2 (P < 0.05). (C and D)The predictive ability of the CpG sites in LINE-1 promoter for TFS(C) and OS(D) by receiver operating characteristic (ROC) curves (*P < 0.001, compared with the other CpG sites).
In addition, a significantly longer median post-resection OS (∼ 60.0 months) was seen in the cluster 1 group compared with 35.0 months (95% CI: 24.0–46.1 months) in the cluster 2 group (log-rank P = 0.0318, Fig.3B). The 1-, 3- and 5-year survival rates in cluster 1 patients, who had higher level of LINE-1 promoter methylation, were 86.11, 61.11 and 52.78% respectively. In contrast, the 1-, 3- and 5-year survival rates in cluster 2 patients, who had a lower level of LINE-1 promoter methylation, were 77.14, 42.86 and 22.86% respectively.
To further determine the association between the methylation level of LINE-1 promoter and the outcomes of post-curative resection, the methylation status of each 17 CpG site with outcomes after curative resection were initially compared with receiver operating characteristic (ROC) curves in the cohort 1. The results showed that the predictive power at CpG site 7 and CpG site 18 was higher than that of the other CpG sites (P < 0.05). The areas under operator curves (AUOCs) for CpG site 7 and CpG site 18 were 0.812 (95% IC: 0.711–0.912) and 0.793 (95% IC: 0.711–0.912) respectively. The prediction of TFS with CpG sites 7 and 8 was significantly greater than that with the other CpG sites (Fig.3C, P < 0.001). The sensitivity and specificity of CpG site 7 for TFS were 77.8 and 80.6% respectively; whereas, the sensitivity and specificity of CpG site 18 were 65.7 and 77.1% respectively. In addition, the prediction value of OS with the AUOC at CpG site 7 and CpG site 18 was significantly greater compared with the others [0.843 (95% IC: 0.755–0.931) for site 7; 0.848 (95% IC: 0.756–0.940) for site 18] (Fig.3D, P < 0.001). The sensitivity and specificity for these two sites were 64, 73.9 and 72, 69.6% respectively. These results strongly suggested that the hypomethylation of CpG sites at 7 and 18 of LINE-1 promoter could be used as a novel potential prognostic biomarker for HCC.
Association of methylation levels of CpG Site7 and CpG site18 with clinical characteristics and outcomes after curative resection
The significant association of the hypomethylation of CpG site 7 and CpG site 18 in cohort 1 with outcomes after curative resection, prompted investigation of the association between the methylation status at CpG site 7 and 18 and clinicopathological parameters in a larger number of patients with HCC, cohort 2. The median follow-up period was 49 months (range: 7–60 months) in cohort 2. The median values of the 7th and the 18th CpG site methylation level were 0.53 (range: 0.1–0.85) and 0.52 (range: 0.1–0.81) respectively. Patients with HCC in cohort 2 were divided into two groups according to the median value of the 7th and the 18th CpG site methylation level. Hypomethylation at the 7th and the 18th CpG sites was significantly associated with larger tumour size, late clinical stage and poor tumour differentiation, but not with gender, age, numbers of tumour or AFP level (Table2).
Table 2.
Comparison of baseline characteristics according to methylation level of site 7 and site 18 in cohort 2
| Variable | Site 7 | Site 18 | ||||
|---|---|---|---|---|---|---|
| Hyper- methylation | Hypo- methylation | P- value | Hyper- methylation | Hypo- methylation | P- value | |
| Age (Mean age ± SD)(y) | 45.71 ± 8.71 | 46.92 ± 9.48 | 0.3835 | 46.17 ± 9.62 | 46.47 ± 8.59 | 0.8306 |
| Gender | ||||||
| Male | 72 | 78 | 0.9181 | 80 | 70 | 0.4699 |
| Female | 13 | 9 | 7 | 15 | ||
| Tumour characteristics | ||||||
| Tumour diameter | ||||||
| <3.8 cm | 63 | 30 | <0.0001 | 62 | 31 | 0.0003 |
| ≥3.8 cm | 22 | 57 | 25 | 54 | ||
| Number of nodules | ||||||
| Single | 74 | 70 | 0.8487 | 78 | 66 | 0.3368 |
| Multinoduar | 11 | 17 | 89 | 19 | ||
| BCLC Stage | ||||||
| Stage A | 56 | 25 | <0.0001 | 57 | 24 | <0.0001 |
| Stage B | 29 | 62 | 30 | 61 | ||
| Tumour differentiation | ||||||
| Well | 26 | 9 | 0.0022 | 24 | 11 | 0.0174 |
| moderate | 34 | 24 | 35 | 23 | ||
| poorly | 25 | 54 | 26 | 51 | ||
| AFP | ||||||
| ≤400 ng/m | 56 | 40 | 0.1409 | 50 | 46 | 0.9955 |
| >400 ng/ml | 29 | 47 | 37 | 39 | ||
Patients with hypermethylation at CpG site 7 had a median OS of ∼ 60 months after receiving curative resection with 1-, 3- and 5-year survival rates of 92.9, 67.1 and 56.5% respectively. However, patients with hypomethylation at CpG site 7 had a median OS of 39 months (95% CI: 26.8–51.2) after receiving curative resection with 1-, 3- and 5-year survival rates of 85.1, 52.9 and 36.8% respectively (P = 0.0048, Fig.4A). Moreover, the median TFS for patients with hypermethylation at CpG site 7 was 43 months (95% CI: 32.4–53.6 months) and 1-, 3- and 5-year TFS were 88.2, 56.5 and 40.0% respectively. These rates were significantly longer than the median TFS of 27 months for patients with hypomethylation site 7 (95% CI: 18.8–35.2 months) and the 1-, 3- and 5-year TFS rates were 78.2, 34.5 and 20.5% respectively (log-rank P = 0.0023; Fig.4B).
Figure 4.
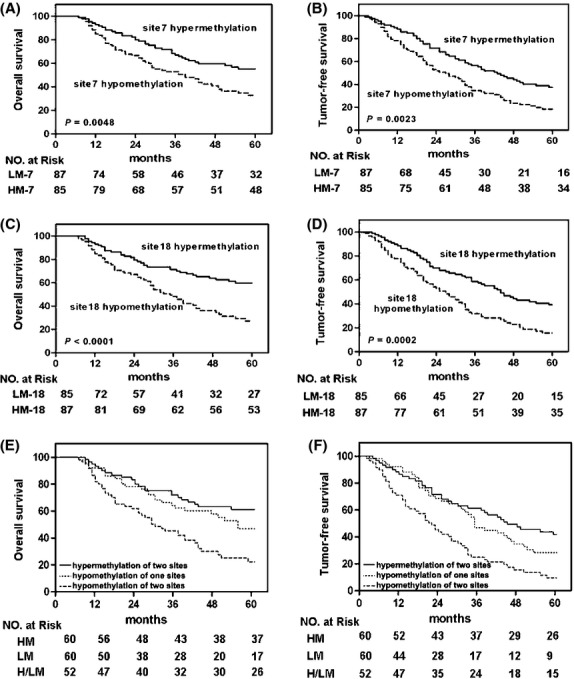
Kaplan–Meier analysis of the correlation between methylation level of CpG site7 or CpG site 18 and outcomes of 172 HCC patients. Patients with CpG site 7 hypermethylation had significantly longer OS (A) or TFS (B) than those with CpG site 7 hypomethylation, and patients with CpG site 18 hypermethylation had significantly longer OS (C) or TFS (D) than those with CpG site 18 hypomethylation. Patients with both CpG site 7 and site 18 hypomethylation had a much shorter median OS (E) or TFS (F) than those with both sites hypermethylation or only one site hypomethylation.
Compared with HCC patients with hypomethylation at CpG site 18,those with hypermethylation at CpG site 18 had a longer median OS (∼60 vs. 35 months respectively, log-rank test, P < 0.0001, Fig.4C) and TFS (44 vs. 27 months; log-rank test, P = 0.0002, Fig.4D). The 1-, 3- and 5-year survival rates for patients with hypermethylated CpG site 18 were 93.1, 71.3 and 60.9% respectively. In contrast, the 1-, 3- and 5-year survival rates for patients with hypomethylated CpG site 18 were 84.7, 48.2 and 31.8% respectively. Furthermore, the 1-, 3- and 5-year TFS rates for patients with HCC and hypermethylated CpG site 18 were 88.5, 58.6 40.2%, respectively, and for those with hypomethylated CpG site 18 were 77.7, 31.8 and 17.7% respectively. Further compared with HCC patients with both CpG site 7 and site 18 hypermethylation or only one CpG site hypomethylation, those with both sites hypomethylation had a shorter median OS (30 vs. ∼60 months and 30 vs. 55 months, respectively, log-rank test P < 0.0001 and P = 0.005, Fig.4E) and TFS (22 vs. 48 months and 22 vs. 35 months, respectively, log-rank test P < 0.0001 and P = 0.0025, Fig.4F). The multivariate analyses showed that the hypomethylation status of CpG site 7 and 18 for patients with HCC after curative resection was independently associated with larger tumour size, poor tumour differentiation, shorter TFS and OS(Table3).
Table 3.
Multivariate analysis of factors associated with survival and recurrence in cohort 2
| Variable | TFS | OS | ||||
|---|---|---|---|---|---|---|
| Univariate P | Multivariate | Univariate P | Multivariate | |||
| HR (95% CI) | P- value | HR (95% CI) | P- value | |||
| Age | 0.965 | 0.488 | ||||
| Gender (male vs. female) | 0.420 | 0.960 | ||||
| Child-Pugh (class A vs. class B) | 0.346 | 0.965 | ||||
| AFP (≤400 ng/ml vs. >400 ng/ml) | 0.454 | 0.638 | ||||
| Tumour diameter (<3.8 cm vs. ≥3.8 cm) | 0.000* | 2.605 (1.313 to 3.247) | 0.002* | 0.000* | 2.228 (1.429 to 3.625) | 0.001* |
| Number of nodules (single vs. multinodular) | 0.035* | 1.149 (0.709 to 1.860) | 0.573 | 0.054 | 1.313 (0.811 to 2.127) | 0.269 |
| BCLC Stage (stage A vs. stage B) | 0.012* | 1.565 (0.958 to 2.559) | 0.074 | 0.005* | 1.622 (1.027 to 2.562) | 0.038* |
| Tumour differentiation (well/mediately vs. poorly) | 0.000* | 0.314 (0.130 to 0.756) | 0.010* | 0.000* | 0.416 (0.277 to 0.627) | 0.021* |
| Site 7 methylation (<0.53 vs. ≥0.53) | 0.001* | 0.435 (0.239 to 0.789) | 0.038* | 0.002* | 0.672 (0.416 to 1.085) | 0.025* |
| Site 18 methylation (<0.52 vs. ≥0.52) | 0.000* | 0.461 (0.256 to 0.832) | 0.010* | 0.001* | 0.550 (0.330 to 0.916) | 0.022* |
Univariate analysis, Cox proportional hazards regression model.
OS, overall survival; TFS, Tumour-free survival.
P < 0.05.
Discussion
It is well documented that the epigenetic alterations play a crucial role in tumourigenesis. LINE-1 promoter methylation is prominent in the genome and is frequently used to assess the status of global methylation 23. In patients with HCC, studies have focused on aberrant methylation of tumour-associated genes 24,25. Recently, LINE-1 hypomethylation has been used as a potential prognostic marker for HCC 26,27. However, the methylation status at each CpG site of the LINE-1 promoter in patients with HBV-related HCC and their clinical significance has not been thoroughly investigated.
In this study, the methylation status of LINE-1 promoter was initially analysed in a pilot cohort of 71 patients with HBV-related HCC, tissue from 29 cases with cirrhosis of the liver and 19 control patients with normal liver tissues. The results showed a significant decrease in the methylation level of LINE-1 promoter in HCC compared with non-tumourous liver tissues. These results are consistent with previous findings that global hypomethylation is a common epigenetic event in HCC compared with cirrhotic and non-cirrhotic tissues 10,28. Interestingly, there were no significant differences in LINE-1 hypomethylation levels among non-HCC samples, suggesting that LINE-1 hypomethylation is one of the specific events associated with tumourigenesis rather than with other pathological processes including HBV infection and cirrhosis. These findings are again consistent with previous studies where levels of hypomethylation in normal liver tissue are comparable with those detected in chronic liver disease 29. The immunohistochemical study also showed that LINE-1 was expressed in cells with strong nuclear staining, but not in non-tumourous hepatocytes. It has been reported that there is asymmetric methylation in the hypermethylated CpG promoter region of the human LINE-1 30. Moreover, methylation of the first seven CpG in LINE-1 promoter is thought to be essential for the inhibition of this gene expression 31. Our results indicate that hypomethylation of the LINE-1 promoter, including site 7 and 18, may result in re-expression of LINE-1 and subsequent translocation into the nuclear 32 as a retrotransposon, leading to chromosomal instability. Thus, hypomethylation of the LINE-1 promoter might play an important role in HCC tumourigenesis, particularly for the progressive and aggressive nature of the disease.
In this study, we found that patients with hypomethylation of the LINE-1 promoter had a significantly shorter median TFS and OS after curative resection compared with those with hypermethylation of the LINE-1 promoter region. Furthermore, hypomethylation of the LINE-1 promoter at CpG site 7 and 18 was significantly associated with poor outcomes after curative resection by ROC curve analysis, with a high degree of sensitivity and specificity. These results suggest that LINE-1 promoter methylation could be used as a potentially prognostic marker in patients with HCC.
These promising initial results prompted us to further investigate LINE-1 methylation as a candidate prognostic indicator in a larger series of patients with HCC. Subgroup analysis of the second cohort of 172 patients with HCC showed that hypomethylation of the CpG site 7 or site 18 revealed a significantly shorter median TFS and OS after curative resection than those with hypermethylation at the CpG site 7 or site 18. More importantly, the median TFS and OS were much shorter in patients with hypomethylation of both CpG site 7 and CpG site 18, which indicate that the combination of these two CpG sites might have significant predictive value in these patients. The hypomethylation of the CpG site 7 or site 18 of cohort 2 in patients with HCC was strongly associated with large tumour size, poor tumour differentiation, worse tumour stage and a higher recurrence rate. The possible consequences of global hypomethylation include chromosomal instability, increase in DNA double-strand breaks, reactivation of transposable elements and activation of antisense transcription 33–36. Chromosomal instability is a hallmark of cancer and is frequently observed in the later stages of tumourigenesis, therefore, the global hypomethylation in patients with HCC might be involved in the tumour progress rather than in the initiation of tumourigenesis. These results are consistent with studies assessing other tumour types where hypomethylation of the LINE-1 promoter is associated with a poor prognosis including prostate adenocarcinoma 37, colon cancer 38, ovarian cancers39 and chronic myeloid leukaemia 40, as well as an association with the aggressiveness of malignant gastrointestinal stromal tumours 41. Interestingly, multivariate analysis showed that the methylation status of the CpG site 7 and 18 of LINE-1 promoter together with tumour size and tumour differentiation could predict outcomes in patients with HBV-related HCC after curative resection. LINE-1 methylation was found to be an independent prognostic factor in cohort 2 patients with HCC. This finding could influence post-operative management of patients following resection, help decide on optimal adjuvant chemotherapy and the frequency of follow-up examination. The prognostic significance of LINE-1 methylation observed in this study should be further validated in a prospective and large-scale clinical study. However, some other well-established prognostic indicators including alpha-fetoprotein and numbers of tumour foci were not associated with post-resection outcomes in this study.
In summary, this study demonstrated that hypomethylation of LINE-1 promoter was associated with tumour progression, larger tumour size, higher recurrence rates, worse tumour stage and poor tumour differentiation in patients with HCC. Moreover, our results strongly suggest that the hypomethylation at CpG site 7 and 18 of LINE-1 promoter region could be used as a potential prognostic biomarker for the prediction of clinical outcomes after curative tumour resection in HBV-related HCC patients and influence future treatment.
Acknowledgments
Financial support: This work was supported by the Key Scientific and Technological Research Foundation of the National Special-purpose Program (2008ZX10002-018), military Special-Purpose Program (No. BWS11J074) and the Capital Medical Research and Development Fund (2009-2041), China.
Conflicts of interest: The authors do not have any disclosures to report.
Glossary
- HCC
Hepatocellular carcinoma
- LINE-1
long interspersed nuclear element-1
References
- 1.Yuen MF, Hou JL, Chutaputti A. Hepatocellular carcinoma in the Asian Pacific region. J Gastroenterol Hepatol. 2009;24:346–53. doi: 10.1111/j.1440-1746.2009.05784.x. [DOI] [PubMed] [Google Scholar]
- 2.Llovert JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–17. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 3.Poon RT, Fan ST, Tsang FH, Wong J. Locoregional therapies for hepatocellular carcinoma: a critical review from the surgeon's perspective. Ann Surg. 2002;235:466–86. doi: 10.1097/00000658-200204000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Burroughs A, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–38. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 5.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–63. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 6.Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol. 2006;21:15–21. doi: 10.1111/j.1440-1746.2005.04043.x. [DOI] [PubMed] [Google Scholar]
- 7.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–38. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 9.Brouha B, Schustak J, Badge RM, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalitchagorn K, Shuangshoti S, Hourpai N, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–6. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 11.Herceg Z, Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat Res. 2011;727:55–61. doi: 10.1016/j.mrrev.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Kim MJ, White-Cross JA, Shen L, Issa JP, Rashid A. Hypomethylation of long interspersed nuclear element-1 in hepatocellular carcinomas. Mod Pathol. 2009;22:442–9. doi: 10.1038/modpathol.2008.203. [DOI] [PubMed] [Google Scholar]
- 13.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 14.Lengauer C, Kinzler KW, Vogelstein B. DNA methylation and genetic instability in colorectal cancer cells. Proc Natl Acad Sci USA. 1997;94:2103–5. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–22. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao XD, Lu YY, Feng F, et al. Effects of LINE -1 ORF-1p over expression on proliferation and anchor- independent growth of SMMC7721 hepatoma cell line. J Clin Hepatol. 2011;14:323–6. [Google Scholar]
- 17.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–3. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 18.Calvisi DF, Simile MM, Ladu S, et al. Altered methionine metabolism and global DNA methylation in liver cancer: relationship with genomic instability and prognosis. Int J Cancer. 2007;121:2410–20. doi: 10.1002/ijc.22940. [DOI] [PubMed] [Google Scholar]
- 19.Chang S, Wang L, Guan Y, et al. Long interspersed nucleotide element-1 hypomethylation in folate-deficient mouse embryonic stem cells. J Cell Biochem. 2013;114:1549–58. doi: 10.1002/jcb.24496. [DOI] [PubMed] [Google Scholar]
- 20.Bruix J, Sherman M, Llovet JM, et al. EASL Panel of Experts on HCC Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 EASL conference. European Association for the Study of the Liver. J Hepatol. 2001;35:421–30. doi: 10.1016/s0168-8278(01)00130-1. [DOI] [PubMed] [Google Scholar]
- 21.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylsitosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.HU MM, Wang Y, Zhang L, et al. Prokaryotic expression of LINE-1 encoded protein L1-ORF1 and preparation of anti-L1-ORF1 antibody. China Biotechnology. 2010;30:7–11. [Google Scholar]
- 23.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang GH, Shim YH, Jung HY, et al. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61:2847–51. [PubMed] [Google Scholar]
- 25.Lee JH, Park SJ, Abraham SC, et al. Frequent CpG island methylation in precursor lesions and early gastric adenocarcinomas. Oncogene. 2004;23:4646–54. doi: 10.1038/sj.onc.1207588. [DOI] [PubMed] [Google Scholar]
- 26.Lin CH, Hsieh SY, Sheen IS, et al. Genome-wide Hypomethylation in Hepatocellular Carcinogenesis. Cancer Res. 2001;61:4238–43. [PubMed] [Google Scholar]
- 27.Tangkijvanich P, Hourpai N, Rattanatanyong P, et al. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta. 2007;379:127–33. doi: 10.1016/j.cca.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 28.Takai D, Yagi Y, Habib N, Sugimura T, Ushijima T. Hypermethylation of LINE-1 retrotransposon in human hepatocellular carcinoma, but not in surrounding liver cirrhosis. Jpn J Clin Oncol. 2000;30:306–9. doi: 10.1093/jjco/hyd079. [DOI] [PubMed] [Google Scholar]
- 29.Shukla R, Upton KR, Munoz-Lopez M, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–11. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodcock DM, Lawler CB, Linsenmeyer ME, Doherty JP, Warren WD. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem. 1997;272:7810–6. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]
- 31.Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–34. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 32.Harris CR, Normart R, Yang Q, et al. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Gene & Cancer. 2010;1:115–24. doi: 10.1177/1947601909360812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 34.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 35.Sassaman DM, Dombroski BA, Moran JV, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 36.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–93. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho NY, Kim BH, Choi M, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–77. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 38.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–8. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pattamadilok J, Huapai N, Rattanatanyong P, et al. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer. 2008;18:711–7. doi: 10.1111/j.1525-1438.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 40.Roman-Gomez J, Jimenez-Velasco A, Agirre X, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–23. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 41.Igarashi S, Suzuki H, Niinuma T, et al. A novel correlation between LINE-1 hypomethylation and the malignancy of gastrointestinal stromal tumors. Clin Cancer Res. 2010;16:5114–23. doi: 10.1158/1078-0432.CCR-10-0581. [DOI] [PubMed] [Google Scholar]