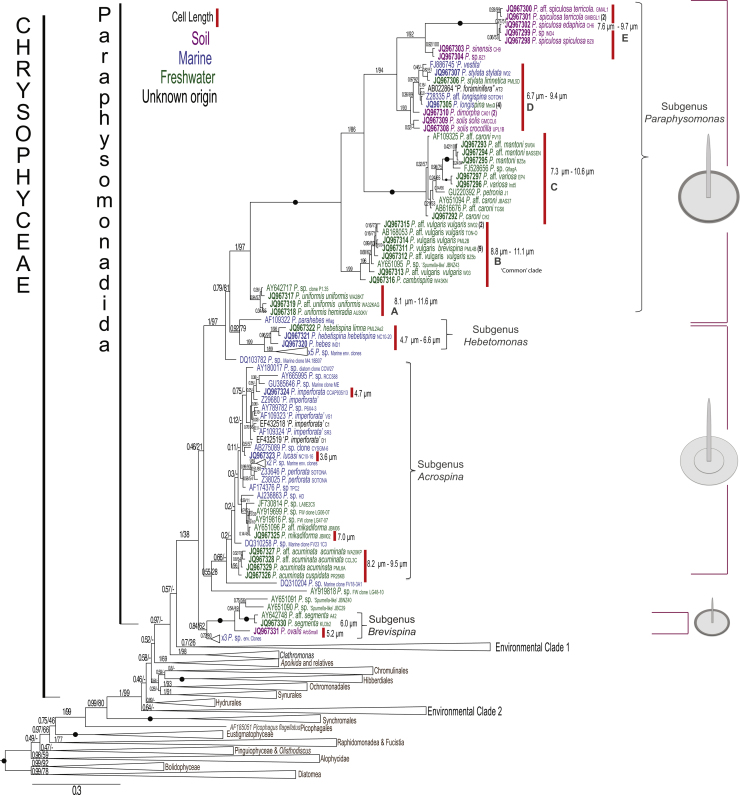
Fig. 1.
MrBayes covarion tree for 329 ochrophyte 18S rDNA sequences showing only the branching order of Paraphysomonas sensu stricto in detail (1672 nucleotide positions). Support values are MrBayes posterior probabilities (left) and RAxML bootstrap percentages for 1000 pseudoreplicates to the right. Black dots mean maximal support for both, i.e. 1/100. All new sequences are in bold type (starting with ‘JQ’). The number of identical sequences obtained in this study from different isolates is shown in parentheses; the common clade included the most commonly found 18S rDNA sequence. The schematic sketches indicate typical scale structure for each of the four subgenera, each corresponding to a single reproducible clade; note how isolates with dense rim to the base-plate of the spine scale group separately from those lacking a prominent rim; scale sizes are arbitrary. The ranges of cell length measurements (from this study only) are indicated beside the red lines. Sequences from freshwater strains are green, from marine strains blue, and soil strains purple. Branching order within the collapsed non-Paraphysomonas chrysophyte taxa are shown in Supplementary Fig. S3, and the outgroups in S2.