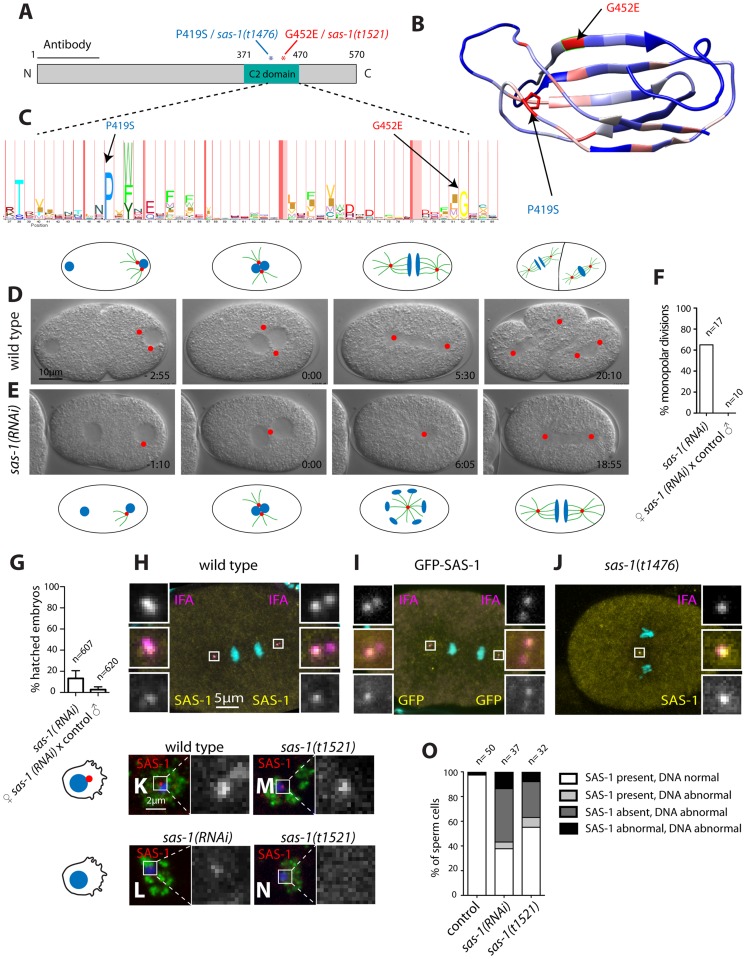
Figure 4. SAS-1 is a C2 domain containing protein that localizes to centrioles.
(A) Schematic of SAS-1 protein architecture with an indication of the single amino acid substitutions in the two sas-1 alleles, and the part against which the antibody was raised. (B) 3D model of the predicted C2 domain of SAS-1 (aa 371–470). Coloring of the residues is according to conservation (blue least, red most). The two amino acids mutated in the sas-1 alleles are indicated. (C) Hidden Markov Model (HMM) representation of the C2 domain, obtained from Pfam (PF00168) [78]. (D–E) Images from DIC time-lapse recordings of wild type (D) and sas-1(RNAi) (E) embryo. (F) Quantification of sas-1(RNAi) phenotype by time-lapse DIC microscopy. (G) Progeny test revealing the paternal requirement for sas-1(RNAi). The two experiments were not from the same batch of RNAi plates, likely explaining the lower viability in the progeny of the mated animals. (H–J) Immunostainings of a GFP-SAS-1 embryo for GFP (yellow) and IFA (magenta) (I), as well as of a wild type (H) or sas-1(t1476) (J) embryo stained for SAS-1 (yellow) and IFA (magenta). DNA is in cyan in all panels. (K–N) Wild type (K), sas-1(RNAi) (L) and sas-1(t1521) (M–N) sperm cells stained with the sperm marker SP-56 (green) and SAS-1 (red and shown alone in magnified panels). DNA is in blue. Note that a signal is present in (M), while it is absent in (N). Note that in (O), SAS-1 abnormal indicates that SAS-1 is either absent or, alternatively, present in too many foci, which we interpret to reflect meiotic defects and/or centrioles in the process of disintegrating. Note also that in sas-1(RNAi), ∼6% of sperm cells do not harbor SAS-6, indicating that under those conditions, some sperm cells may be without centrioles altogether. Note that these experiments were performed with sas-1(t1521). (O) Quantification of experiments shown in (K–N). See also Fig. S6.