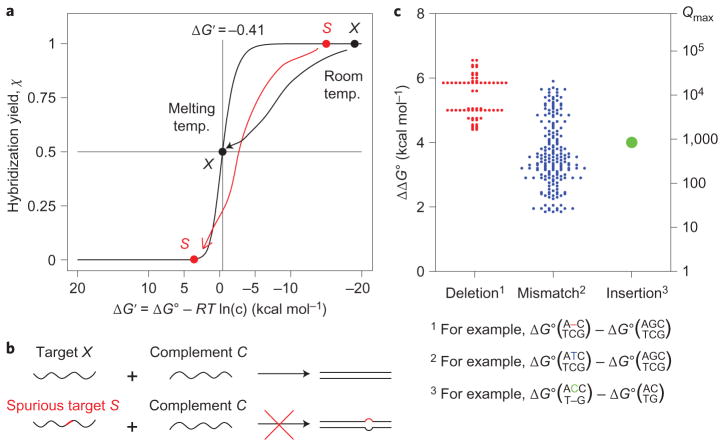
Figure 1. Hybridization specificity of nucleic acids.
a, Hybridization yield χ plotted against concentration-adjusted standard free energy ΔG′ = ΔG∘ + (Δn)RTln(c), where c is the concentration of the limiting species, and Δn = −1 for a standard bimolecular hybridization reaction. At room temperature, the binding of both the correct target (black dot) and the spurious target (red dot) are thermodynamically favourable and practically indistinguishable. In contrast, at the melting temperature, ΔG′ = −RTln((c/2)/(c/2)2)−RTln(c) = −0.41 kcal mol−1, the hybridization yield of the correct target is 50%, and much lower for the spurious target. b, In a hybridization-based assay or reaction, specificity is achieved when a spurious target that differs in sequence from the correct target by a single base (depicted as the red segment) does not hybridize significantly to the complement. c, The standard free energy difference (ΔΔG∘) caused by a single-base change ranges from 1.83 to 6.57 kcal mol−1, and determines the upper bound on the discrimination factor: Qmax ≡ eΔΔG∘/RT. (Graphic constructed using thermodynamic parameters by SantaLucia and Hicks22; see Supplementary Text S3 and Tables S1–S5 for detailed numerical values.) All 64 cases of single-base insertion were modelled to have identical ΔΔG∘.