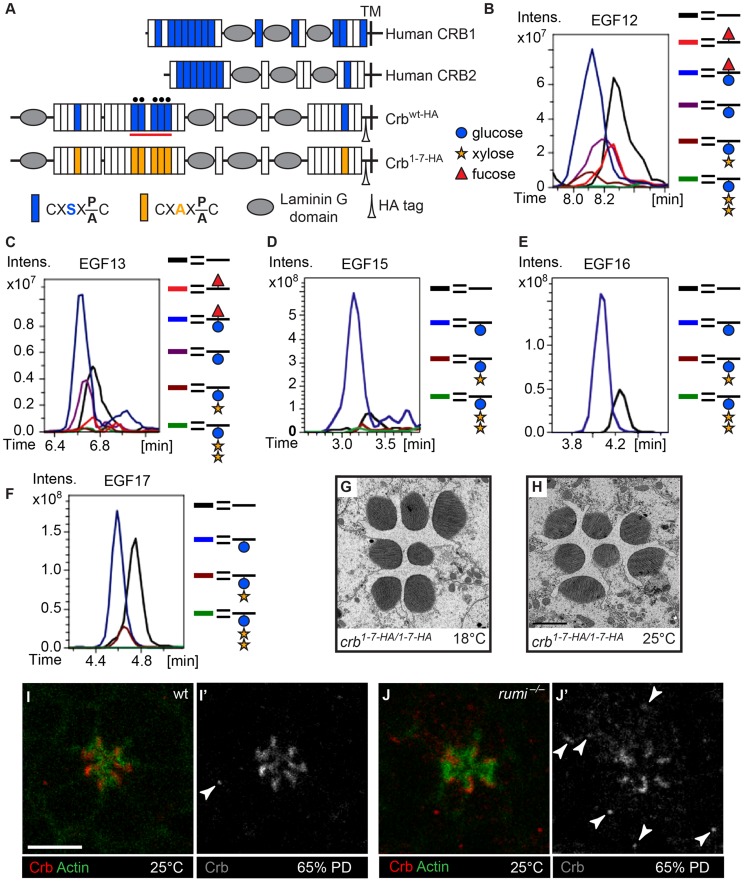
Figure 3. Loss of O-glucose on Crb cannot explain the rumi−/− rhabdomere attachment phenotype.
(A) Schematic of the human CRB1, human CRB2, and wt and mutant, HA-tagged Drosophila Crb based on the Crb-PA polypeptide (FlyBase ID: FBpp0083987). Blue and orange boxes indicate EGF repeats harboring wt and mutant Rumi consensus sequences, respectively. TM, transmembrane domain. Black circles mark EGF repeats shown in B–F. (B–F) Extracted Ion Chromatogram (EIC) data for peptides containing the O-glucose consensus site from Crb EGF12 (B), EGF13 (C), EGF15 (D), EGF16 (E) and EGF17 (F) obtained from mass spectrometry on a Crb fragment indicated by the red line in (A). Note the presence of O-glucose (blue circle) on all five EGF repeats, some of which are elongated by xylose (yellow star). Peptides from EGF12 and EGF13 also harbor O-fucose (red triangle) added to the consensus O-fucosylation sites present in these two EGF repeats. Full spectra and the peptide sequences are shown in Figure S2 and Figure S3. (G,H) crb knock-in mutants lacking all Rumi target sites and raised at 18°C (G) or 25°C (H) do not show rhabdomere attachment. Scale bar in H applies to G and H and is 2 µm. (I–J′) Loss of Rumi does not impair Crb localization to the stalk membrane. Increased Crb puncta, some of which are marked with arrowheads in J′, are seen in the photoreceptor cell body of rumi−/− ommatidia raised at 25°C (J,J′) compared to wild-type (I,I′). Actin (green) is used to mark the rhabdomeres. Crb is shown in red. Scale bar: 5 µm.