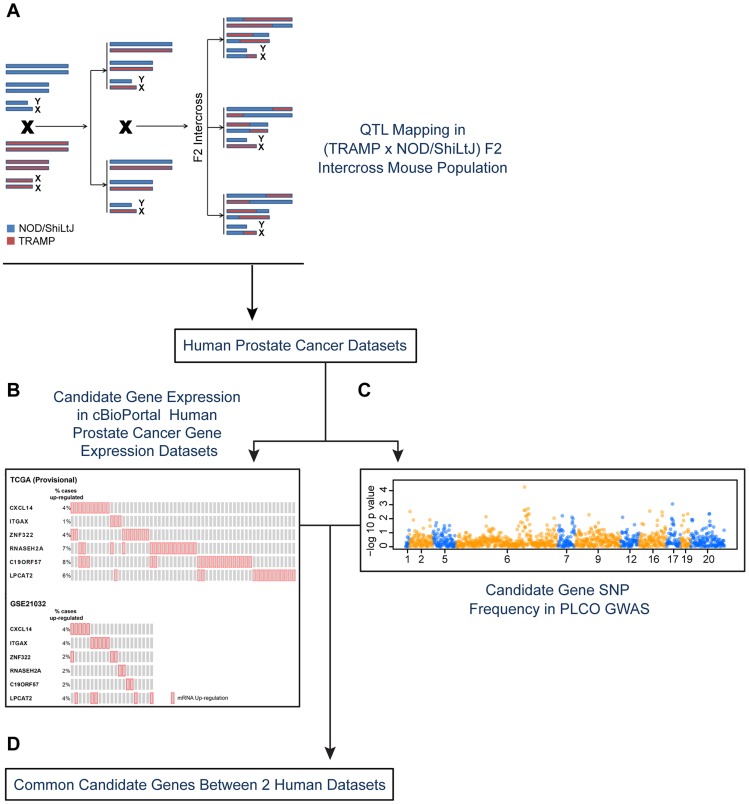
Figure 1. Experimental strategy for identifying novel susceptibility genes for aggressive prostate cancer.
Candidate aggressive disease modifier genes were identified in an F2 intercross population involving the TRAMP mouse model of prostate tumorigenesis and the NOD/ShiLtJ strain of mouse, which is highly susceptible to aggressive disease development (A). This strategy involved QTL mapping to identify genomic regions associated with aggressive disease traits, followed by eQTL mapping and gene expression-trait correlation analyses to nominate candidate modifiers. Following this, a strategy involving two types of data derived from human prostate patients was used to nominate the highest priority candidate genes: (B) human prostate cancer primary tumor gene expression datasets; and (C) a human prostate cancer GWAS dataset. Only those genes designated as being associated with aggressive disease development in both the tumor gene expression and GWAS datasets were designated as being high priority candidate genes (D).