Abstract
Background
Leprosy is characterized by polar clinical, histologic and immunological presentations. Previous immunologic studies of leprosy polarity were limited by the repertoire of cytokines known at the time.
Methodology
We used a candidate gene approach to measure mRNA levels in skin biopsies from leprosy lesions. mRNA from 24 chemokines and cytokines, and 6 immune cell type markers were measured from 85 Nepalese leprosy subjects. Selected findings were confirmed with immunohistochemistry.
Principal Results
Expression of three soluble mediators (CCL18, CCL17 and IL-10) and one macrophage cell type marker (CD14) was significantly elevated in lepromatous (CCL18, IL-10 and CD14) or tuberculoid (CCL17) lesions. Higher CCL18 protein expression by immunohistochemistry and a trend in increased serum CCL18 in lepromatous lesions was observed. No cytokines were associated with erythema nodosum leprosum or Type I reversal reaction following multiple comparison correction. Hierarchical clustering suggested that CCL18 was correlated with cell markers CD209 and CD14, while neither CCL17 nor CCL18 were highly correlated with classical TH1 and TH2 cytokines.
Conclusions
Our findings suggest that CCL17 and CCL18 dermal expression is associated with leprosy polarity.
Author Summary
Leprosy presents with a polarized spectrum, with lepromatous leprosy having high bacillary numbers and TH2 dermal cytokines, versus tuberculoid leprosy showing very few bacilli and TH1 cytokines. The mechanism underlying this polarized presentation is largely unknown. In the following study, we isolated mRNA from skin biopsies from 85 individuals with leprosy and measured the expression of a panel of 24 cytokines and 6 cell markers. We found that three soluble mediators (CCL17, CCL18 and IL10) and one cell marker (CD14) were differentially expressed in leprosy dermal lesions. CCL18 and IL10 were more highly expressed within lepromatous lesions, and CCL17 and CD14 were more highly expressed within tuberculoid lesions. In addition, CCL18 protein expression was confirmed by immunostaining. CCL17 and CCL18, were more strongly associated with leprosy polarity than traditional TH1 and TH2 cytokines. These data suggest that newer soluble chemokines may be important in leprosy pathogenesis and uncover a molecular signature of the two polar phenotypes of leprosy, which may be useful in future diagnostics.
Introduction
Leprosy is a spectrum of illnesses involving the nerves, skin and upper airways of humans [1]. Depending on the host response to Mycobacterium leprae, the clinical presentation can vary widely between individuals [2]. Histologic features of these lesions can be quantitated on a 5 point classification system (Tuberculoid tuberculoid (TT), Borderline Tuberculoid (BT), Borderline Borderline (BB), Borderline lepromatous (BL), and Lepromatous Lepromatous (LL)) defined by Ridley and Jopling in 1966 which helps to define the polar spectrum found in clinical disease cause by M leprae [3]. In lepromatous leprosy (LL and BL), the clinical course and histologic features are distinguished by uncontrolled replication of the bacilli in dermal foamy macrophages and poor granuloma formation. In tuberculoid leprosy (BT and TT), the skin lesions are characterized by well-demarcated granulomatous inflammation within nerves and skin, CD4 and CD8 T cell dermal infiltration, and little evidence of bacilli within lesions.
Early studies of leprosy intradermal cytokine expression determined that leprosy polarity is associated with TH2 cytokines IL4, IL5, IL10 in lepromatous lesions, and TH1 polar cytokines, IFNG, IL2, in tuberculoid lesions [4]. In addition, CD4 T-cell frequency was increased in the tuberculoid lesions compared to the lepromatous lesions [5]. Since these early seminal studies, many additional cytokines and chemokines have been identified but not analyzed in leprosy lesions. Genome-wide transcriptional profiling has also been used to define cytokine mRNA levels within lesions [6]. The sample size of polar subgroups was low in this study and precluded a full comparison of the tuberculoid and lepromatous groups. To date, a detailed study of leprosy using larger sample sizes and surveying a broad, current repertoire of cytokines and chemokines has not been performed.
Differentiation and polarization of T cell subsets is characterized by a complex network of cytokines and chemokines [7], [8]. Initially, T-cell subsets were defined by their release of specific cytokines; TH1 associated with IFNG and IL2 and TH2 subset associated with release of IL4, IL5, and IL10. Since these influential studies, the number of cytokines that defined T cell subsets have been expanded to include TH17, TH22, and Tregs [8]. CCL17 and CCL18 are chemokines important in T cell mediated reactions [9]–[15]. CCL17 is secreted by alternatively activated macrophages [16] and is markedly elevated in patients with allergic atopic dermatitis who have a TH2 dominance [15]. In murine studies, CCL17 has been implicated in both TH1 and TH2 responses [15]–[19]. CCL17 is increased in dendritic cells activated with M. tuberculosis [20]. CCL18, is elevated in atopic dermatitis [21] and also is secreted by dendritic cells upon recognition of MTB by innate immune cells [22]. Additionally, CCL18 has been implicated in differentiation of macrophages into an alternatively activated phenotype [23].
In the following study, we measured the mRNA levels of 24 soluble cytokines and 6 cell specific markers in 82 individuals with biopsy-confirmed polar leprosy with the goal to better characterize and define the immune responses in individuals with leprosy phenotypes. Herein we show that two chemokines, CCL17 and CCL18, more accurately define leprosy polarity than traditional TH1 and TH2 cytokines.
Methods
Ethics Statement
All human subjects provided informed consent to the procedures below. No children were enrolled in the study. Both written and oral informed consent was given due to the high rates of illiteracy. Subjects that were unable to read and write provided a thumbprint as proof of consent, while those that could read and write provided a signature. All informed consent documents and procedures were approved by the University of Washington Institutional Review Boards per the US Department of Health and Human Services Guidelines, The Medical Ethics Committee of Leiden University Medical Center, and Nepal Health Research Council (NHRC).
Human Subjects and Study Design
Dermal biopsies were obtained from patients at Anandaban Hospital in Kathmandu, Nepal. These cases comprised more than 8 different ethnic and religious groups included Vaishya, Chhetri, Brahmin, and Sudra (Table 1). Eighty five patients were enrolled based on clinical presentation with the leprosy diagnosis confirmed by skin slit smears and biopsy. One of the biopsy samples was used for histological diagnosis and the other for isolation of mRNA. Leprosy class was determined by Ridley Jopling (RJ) classification [3] after the biopsies were fixed, mounted in paraffin, and stained with both Fite stain and hematoxylin and eosin for viewing under conventional light microscopy by experienced leprosy pathologists at the Schieffelin Institute of Health – Research & Leprosy Centre, Karigiri, Tamil Nadu, India. mRNA levels from dermal biopsies were measured by RT-PCR with a BioMark Fluidigm platform which included 38 with tuberculoid (borderline tuberculoid (BT) and tuberculoid (TT)), 3 with borderline borderline (BB), and 44 with lepromatous leprosy (borderline lepromatous (BL) and lepromatous (LL)) (Table 1). Although Bacillus Calmette–Guérin (BCG) vaccination for the participants was not recorded, it is routine for individuals in Nepal to get vaccinated as an infant in Nepal. Routine population-wide vaccination began in 1966 and therefore individuals born before then may have not received the BCG vaccine as an infant [24]. The biopsies were obtained in a leprosy tertiary care center, where a significant number of people diagnosed with leprosy reactive states are referred. 48 patients out of 85 undergoing analysis for mRNA levels had reactions at the time of biopsy, including the three diagnosed with BB disease. 45 of these patients were included in the polarity analysis and shown in Table 1. For ELISA studies 20 additional, newly diagnosed patients and 6 control individuals from the same region (endemic controls: EC) had serum collected. In addition, skin biopsies were performed as above to determine RJ classification.
Table 1. Leprosy biopsy demographics.
| Tuberculoid | Lepromatous | pvalue | ||||
| Age | (Mean±SD) | 35.7 | ±15.6 | 37.3 | ±14.4 | 0.65 |
| Sex | male (%) | 24 | (63%) | 39 | (89%) | |
| female (%) | 14 | (37%) | 5 | (11%) | 0.01 | |
| Ethnicity | Vaishya | 8 | (21%) | 17 | (39%) | |
| Chhetri | 8 | (21%) | 7 | (16%) | ||
| Other | 8 | (21%) | 7 | (16%) | ||
| Brahmin | 7 | (18%) | 7 | (16%) | ||
| Sudra | 7 | (18%) | 5 | (11%) | ||
| Indian | 0 | (0%) | 1 | (2%) | 0.51 | |
| Reaction | RR1 | 16 | (42%) | 20 | (45%) | 0.76 |
| ENL2 | 0 | (0%) | 9 | (20%) | <0.01 | |
Reversal Reaction,
Erythema nodosum leprosum.
RNA Isolation and RT-PCR
Half of each biopsy sample was macerated in RNAlater (Invitrogen, Carlsbad, CA) preservative and stored at −20°C for later processing. Samples were homogenized with a high shear homogenizer (OMNI international, Kennesaw GA) using disposable tips. Total RNA was isolated using RNeasy mini columns and cDNA was made using Applied Biosystems high capacity cDNA reverse transcriptase kits (Foster City CA). RT-PCR was performed with PrimeTime primer probe sets from Integrated DNA Technologies (Corallville, IA) (Table 1) and Taqman (Life Technologies) for genes CD1a (catalog number Hs00233332_m1) using the Fluidigm 48×48 dynamic array platform. Briefly 2.5 ng of DNA was preamplified in a 5 ul reaction with 25 nM concentration of all primers and 12.5 nM concentration of all probes (For list see Supplemental Table S1) and amplified for 15 cycles with a 30 sec denaturation step at 98°C and 4 minutes at 60°C. The pre-amplified reaction was added to the Fluidigm 48×48 dynamic array platform for each individual diluted in 2× mastermix buffer. 10× IDT PrimeTime gene expression assays were added to the assay portion of the Fluidigm chip and amplified for an additional 40× cycles in the Fluidigm assay chip. To verify that the Fluidigm assay was accurate, for a set of probes (CCL17, CCL18, and CD1a) we did single 20 ul RT PCR assays. Both singleplex assays and Fluidigm chip assays values were corrected by the GAPDH values to control for variability in biopsy size, mRNA yield, and cellularity within biopsy samples. Single assay RTPCR and Fluidigm values had R2 values of approximately 0.7–0.9 values (Supplemental Figure S1).
Immunohistochemistry
4 um paraffin sections were deparaffinized and rehydrated with heat-mediated antigen retrieval performed in citrate buffer (pH 6). Slides were blocked with 2.5% normal horse serum, incubated with anti-CCL18 primary antibody (Peprotech, Catalog # 500-P108) and anti-CCL17 (RND Systems, Catalog #AF364) overnight at 4°C followed by ImmPress rabbit HRP (CCL18) and ImmPress goat HRP (CCL17) (Vector Laboratories, Burlingame CA). Slides were developed with QuantoDAB (Fisher Scientific) and counter-stained with hematoxylin. Stained tissue biopsies were scored by a dermatologist blinded to the subject's leprosy classification. Sections were surveyed on low power, and representative areas showing dermal or subcutaneous inflammation on each specimen were identified. Several representative 40× fields were assessed in each specimen and scored as 0 for <1%, 1+ for 1–10%, 2+ for 10–20%, or 3+ for >20% cells staining for CCL18.
ELISA. Six (6) control and 20 individuals with documented leprosy had sera collected and assayed for CCL17 and CCL18 protein levels by sandwich ELISA (RND biosystems), per manufacturers protocol.
Statistics
The Mann-Whitney U-test was used to make comparisons of the cytokine production between groups, as small sample sizes precluded an assumption of normal distribution. Two-sided testing was used for all comparisons to evaluate statistical significance. A P value of ≤0.05 was considered significant in initial analysis. Bonferroni corrections for multiple comparisons were added as described. Statistics were calculated with Stata software (version 11.2). For dermal expression correlation the non-parametric Spearman's rank correlation test rho (ρ) statistic was used for correlation coefficient, and was generated using R program version 3.0.1 (R: A Language and Environment for Statistical Computing, Vienna Austria). For iterative analysis, we used Stata 11.0 random number generator to randomly assign discovery and validation cohorts on 40 successive iterations and used Mann-Whitney U test for significance. For iterative analysis P values were not adjusted for multiple comparisons; P values of less than 0.05 were considered significant. Hierarchical clustering of the Spearman's ρ statistics was performed in R program using the complete-linkage method [25] which clusters individual tests based on the maximum distance between tests [26]. Graphical representation of clustering was displayed using corrplot package by Teiyun Wei in R program [27], [28].
Accession Numbers (RefSeq)
The following HGNC genes and corresponding refseq accession numbers were used in the paper: CCL1, NM_002981; CCL17, NM_002987; CCL18, NM_002988; CCL2, NM_002982; CD14, NM_000591; CD1A, NM_001763; CD209, NM_021155; CD22, NM_001771; CD3D, NM_000732; FOXP3, NM_014009; GAPDH, NM_002046; IFNA1, NM_024013; IFNA8, NM_002170; IFNB1, NM_002176, IFNG, NM_000619; IL10, NM_000572; IL12A NM_000882; IL12B NM_002187; IL13, NM_002188; IL17A, NM_002190; IL18, NM_001562; IL1B, NM_000576; IL1RN, NM_000577; IL21, NM_021803; IL22, NM_020525; IL23A, NM_016584; IL27, NM_145659; IL4, NM_000589; IL6, NM_000600; TNF, NM_000594; IFNL1, NM_172140
Results
Cytokine/Chemokine Expression in Dermal Lesions from Leprosy Patients
We examined whether skin mRNA levels of candidate immune genes differed in individuals with tuberculoid (TT/BT, n = 38) and lepromatous (BL/LL, n = 44) leprosy. The cohort included Nepalese adults evaluated at a tertiary care center for leprosy (Table 1). Skin biopsies were analyzed for mRNA levels of TH1 and TH2 chemokines/cytokines, type I, II and III interferons, other T-cell associated cytokines, IL12 family members, IL1β family members, cellular markers, and house-keeping genes (for complete list see Table S1). Of the 24 soluble mediators of inflammation that were measured, 7 showed significant differences between lepromatous and tuberculoid leprosy (CCL2, CCL17, CCL18, IFNA1, IL10, IL22, and TNF) (Table 2). CCL2, CCL18, IL10 expression was higher in lepromatous lesions, while CCL17, IFNA1, TNF, and IL22 were more highly expressed in tuberculoid lesions. In addition, median values for the prototypic TH1 cytokine, IFNG, were higher in tuberculoid lesions compared to lepromatous, but it was not statistically significant (median values = 0.212 vs 0.015, P = 0.103). Only CCL17, CCL18 and IL-10 remained significant after adjustment for multiple comparisons using Bonferroni correction. Stratification of samples by type 1 reaction for leprosy reactive state did not alter the associations observed for CCL17 and CCL18 (For P values see below). In order to adjust for age we analyzed CCL17 and CCL18 associations with leprosy phenotype in patients born before 1966 (cutoff age of 45). We found that the majority of the association of CCL17 and CCL18 expression was seen in individuals less than 45 (age >45 n = 26: CCL17, P = 0.09; CCL18, P = 1.4×10−3), (age <45, n = 56; CCL17, P = 8.2×10−4; CCL18, P = 5.7×10−5).
Table 2. Cytokine expression normalized by GAPDH in dermal leprosy lesions.
| TT+BT | LL+BL | |||||
| Cytokine | median | 75 (%) | median | 75 (%) | pValue | pValue (adj) |
| CCL1 | 4.50E-05 | 5.20E-04 | 0.000 | 1.41E-04 | 0.212 | 5.081 |
| CCL2* | 0.033 | 0.161 | 0.146 | 0.372 | 0.009 | 0.218 |
| CCL17 | 0.024 | 0.067 | 0.005 | 0.011 | 2.36E-04 | 0.006 |
| CCL18 | 0.041 | 0.089 | 0.309 | 0.493 | 3.00E-07 | 7.20E-06 |
| IFNA1* | 0.009 | 0.013 | 0.006 | 0.009 | 0.008 | 0.182 |
| IFNA8 | 0.045 | 0.059 | 0.034 | 0.064 | 0.503 | 12.067 |
| IFNB1 | 0.052 | 0.067 | 0.040 | 0.072 | 0.357 | 8.571 |
| IFNG | 0.212 | 0.479 | 0.015 | 0.228 | 0.103 | 2.478 |
| IL10 | 0.011 | 0.028 | 0.044 | 0.085 | 0.001 | 0.022 |
| IL12a | 0.000 | 1.22E-04 | 0.000 | 2.28E-04 | 0.664 | 15.928 |
| IL12b | 0.001 | 0.003 | 0.001 | 0.003 | 0.615 | 14.758 |
| IL13 | 0.000 | 0.000 | 0.000 | 0.000 | 0.326 | 7.831 |
| IL17a | 0.000 | 1.73E-04 | 0.000 | 5.10E-05 | 0.275 | 6.612 |
| IL18 | 0.092 | 0.130 | 0.078 | 0.104 | 0.204 | 4.904 |
| IL1b | 0.022 | 0.048 | 0.018 | 0.040 | 0.270 | 6.481 |
| IL1ra | 2.497 | 3.804 | 1.766 | 3.385 | 0.787 | 18.898 |
| IL21 | 0.000 | 2.36E-04 | 0.000 | 1.26E-04 | 0.645 | 15.470 |
| IL22* | 0.000 | 5.05E-06 | 0.000 | 0.000 | 0.047 | 1.129 |
| IL23 | 0.011 | 0.025 | 0.007 | 0.014 | 0.091 | 2.193 |
| IL27 | 0.000 | 4.96E-05 | 0.000 | 1.13E-04 | 0.729 | 17.494 |
| IL29 (IFNL1) | 0.000 | 0.000 | 0.000 | 7.42E-06 | 0.697 | 16.739 |
| IL4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.484 | 11.611 |
| IL6 | 0.006 | 0.015 | 0.009 | 0.018 | 0.461 | 11.069 |
| TNF* | 0.032 | 0.047 | 0.023 | 0.032 | 0.028 | 0.675 |
TT+BT is tuberculoid polar leprosy, LL+BL is lepromatous polar leprosy. Values with Ct Values greater than 40 were given a value of zero and included in the statistical analysis to determine medians and 75th percentile values. P values were calculated after Mann-Whitney analysis. Adjusted p values are the original p values corrected for multiple comparisons by traditional Bonferroni correction.
In bold are cytokines that were significant following Bonferroni correction.
*Represents cytokines that were significant by Mann-Whitney, but failed multiple comparisons corrections.
To further adjust for multiple comparisons, we randomly arranged our data into a discovery (n≈20) and validation cohort (n≈20) and ran the analyses on 40 separate sample iterations. Increased CCL18 levels were associated with lepromatous leprosy in randomly generated discovery and validation cohorts in 40/40 (100%) of the iterations, while increased CCL17 levels were associated with tuberculoid lesions in 29/40 (73%). On the other hand, IL10 expression, only distinguished polarity in 18/40 (45%) of the iterations.
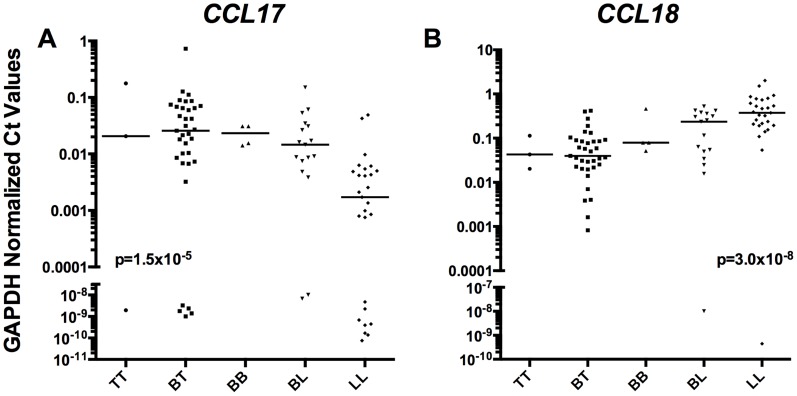
Both CCL17 (Figure 1A) and CCL18 (Figure 1B) expression were highly significant when analyzed in rank order based on Ridley-Jopling classification (non-parametric trend test, P = 1.5×10−5 and 3.0×10−8, respectively). Together, these data suggest that both CCL17 and CCL18 dermal mRNA levels are associated with tuberculoid and lepromatous leprosy, respectively and are more strongly associated with polarity than common TH1/TH2 markers typically used to characterize leprosy lesions.
Figure 1. CCL17 and CCL18 by Ridley Jopling classification.
Relative tissue mRNA levels of CCL17 (A) and CCL18 (B) normalized to GAPDH were arranged in order of specific leprosy classification and analyzed for significance by non-parametric trend test with variables TT, BT, BB, BL, and LL treated as increasing categorical variables, p values displayed. Lines are medians. A value of 40 was given to undetectable Ct Values in order to graphically represent on log scale.
Cellular Marker Expression in Leprosy Skin Biopsies
Next we analyzed the lesions for mRNA expression of markers known to distinguish innate immune cells from B and T cells (CD14-macrophage, CD209-dendritic cell, CD1a-dendritic and Langerhans cells, CD22-B cell, CD3d-T cells, FoxP3-T-Regulatory cells) (Table 3). We measured cell specific mRNA to both determine which cell types were predominant in the lesions and whether certain mediators were associated with a particular cell type. CD14 and CD209 were more highly expressed in lepromatous lesions, while CD1a had higher expression in tuberculoid lesions. CD14 remained significant after corrections for multiple comparisons with Bonferroni adjustment. These data confirm that macrophage infiltration is a characteristic of lepromatous lesions.
Table 3. Cell marker expression in dermal leprosy lesions.
| TT+BT1 | LL+BL2 | |||||
| Cytokine | median | 75 (%) | median | 75 (%) | pValue3 | pValue (adj)4 |
| CD14 | 4.50E-05 | 5.20E-04 | 0.000 | 1.41E-04 | 1.57E-04 | 0.001 |
| CD1a* | 0.033 | 0.161 | 0.146 | 0.372 | 0.011 | 0.067 |
| CD209* | 0.024 | 0.067 | 0.005 | 0.011 | 0.031 | 0.186 |
| CD22 | 0.041 | 0.089 | 0.309 | 0.493 | 0.748 | 4.490 |
| CD3d | 0.009 | 0.013 | 0.006 | 0.009 | 0.088 | 0.528 |
| FoxP3 | 0.045 | 0.059 | 0.034 | 0.064 | 0.214 | 1.282 |
Tuberculoid polar leprosy,
lepromatous polar leprosy. Values of 0.000 are Ct values less than 1/240.
P values were calculated by Mann-Whitney tests. Both median and 75th percentile (75(%)) are depicted.
Adjusted p values are the original p values corrected for multiple comparisons by traditional Bonferroni correction.
In bold are cytokines that were significant following Bonferroni correction.
*represents cytokines that were significant by Mann-Whitney, but failed multiple comparisons corrections.
Analysis of Cytokine and Cell Marker Expression in Leprosy Reactive States
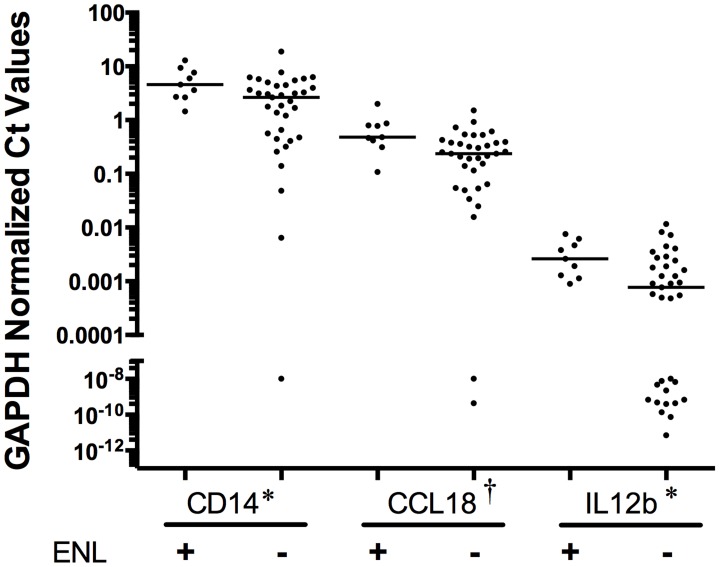
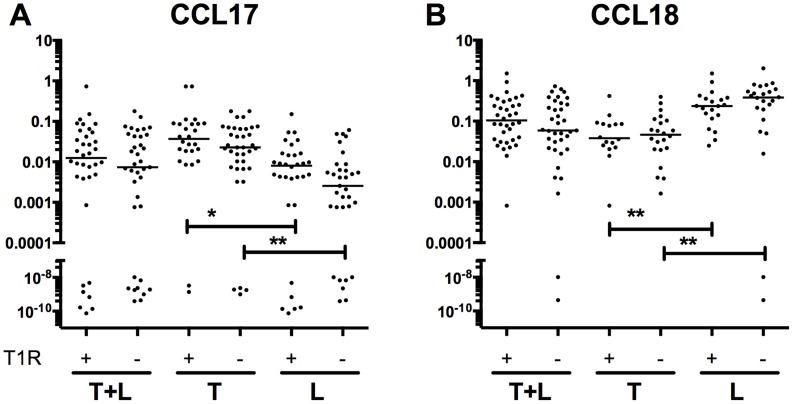
We next examined whether cytokine and cellular markers were significantly different between clinical leprosy reactive states. We analyzed expression levels in BL/LL patients with type 2 ENL reactions (n = 9) compared to those without type 2 ENL reactions (n = 35) and determined that transcripts from mediators CCL18, IL12b, and cell marker CD14 were significantly elevated in lesion samples with ENL (Figure 2). However, all of these failed to reach significance when adjusted for multiple comparisons. Next we examined mRNA levels in patients from whom biopsies were obtained with ongoing type 1 reversal reaction (n = 36, 42% of the tuberculoid patients and 45% of lepromatous patients) versus those who had no reversal reaction (n = 46, 58% of tuberculoid patients and 55% of lepromatous patients). No marker distinguished these two clinical phenotypes. In addition we performed subgroup analysis of mRNA level differences between patients with type 1 reversal reaction and those without type 1 reversal reaction limited to individuals with either tuberculoid leprosy or lepromatous leprosy, and also saw no differences in the cytokines transcript levels examined. Cytokine levels of CCL17 (Figure 3A) and CCL18 (Figure 3B) were significantly different between lepromatous and tuberculoid lesions regardless of the type 1 reactive state (CCL17 with T1R P = 0.0038, without T1R P = 0.011, CCL18 with T1R p = 3.6×10−4, without T1R P = 2.6×10−4).
Figure 2. Dermal levels of CD14, CCL18, and IL12a mRNA in patients with ENL compared to those without.
Expression levels of CD14, CCL18, and IL12a (normalized by GAPDH expression) were compared in patients with ENL (n = 9) versus those in the lepromatous pole without ENL (n = 35). Lines are medians. A value of 40 was given to undetectable Ct values in order to graphically represent on log scale. P values were †<0.01, *<0.05, based on Mann-Whitney analysis.
Figure 3. CCL17 and CCL18 mRNA are not associated with reactive state.
CCL17 and CCL18 mRNA levels (normalized by GAPDH expression) were compared in LL+BL versus TT+BT patients with T1R (L pole (n = 16), T pole (n = 20)) versus those without T1R (L pole (n = 17), T pole (n = 22)). Lines are medians. A value of 40 was given to undetectable Ct Values in order to graphically represent on log scale. P values were **<0.01, *<0.05, based on Mann-Whitney analysis.
Increased CCL18 Protein Staining in Lepromatous Lesions
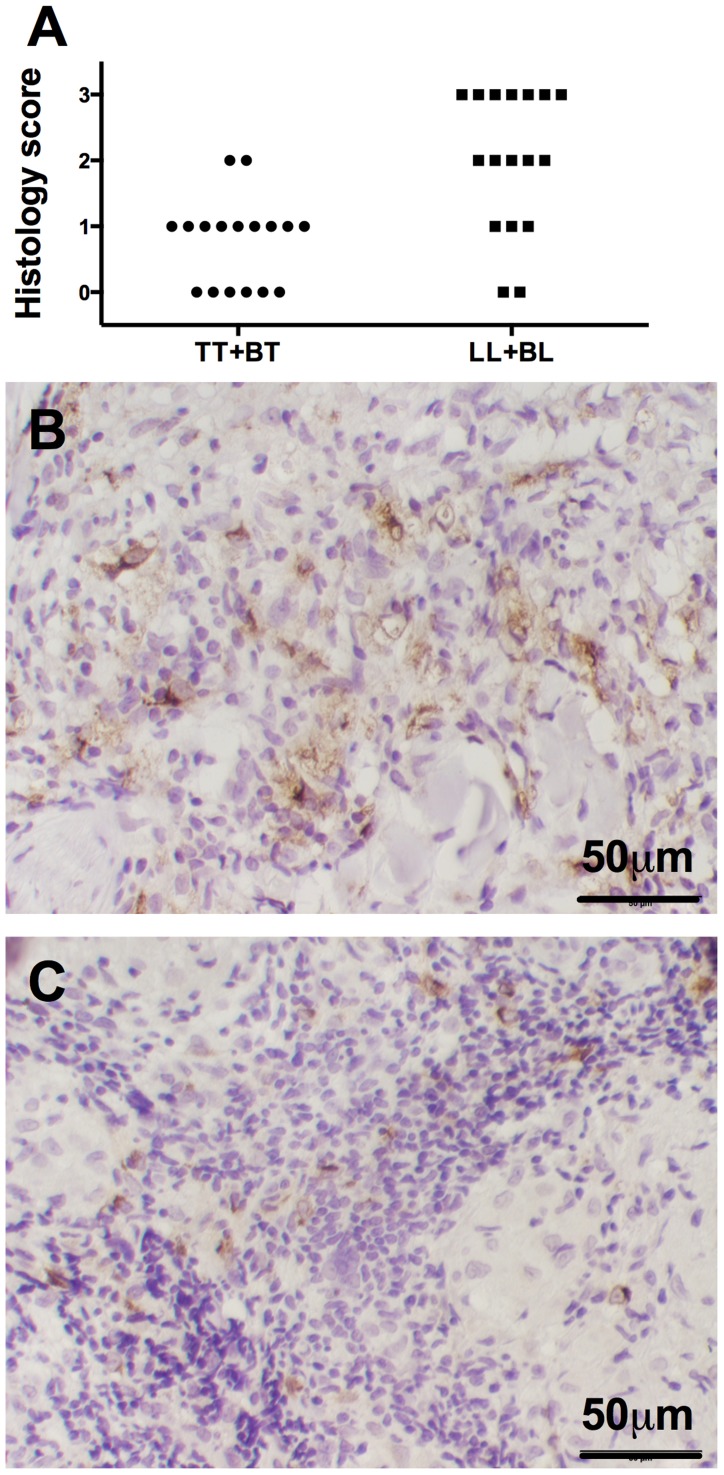
Next we examined CCL17 and CCL18 protein expression within dermal lesions (n = 17 LL+BL and 17 TT+BT) using antibodies directed to human CCL17 and CCL18. Control staining using secondary antibodies showed no immunoreactivity. We were unable to visualize CCL17 staining above background levels despite using multiple antibody dilutions. CCL18 stained cells were detected, primarily in the cytoplasm of large polygonal cells within the dermis and subcutaneous tissues most consistent with the monocyte-derived histiocytes typically found in such inflammation. Lesions stained with CCL18 antibodies were scored semi-quantitatively for percent of immunoreactive cells (Figure 4A). More CCL18 staining was seen in LL+BL lesions (Figure 4B) compared to TT+BT lesions (Figure 4C) (non-parametric Mann-Whitney test P<0.01).
Figure 4. CCL18 immunohistochemistry in leprosy dermal biopsies.
A) Semi-quantitative score of CCL18 immunohistochemical staining in LL+BL (n = 17) and TT+BT (n = 17) lesions. P<0.01 by non-parametric Mann-Whitney analysis. Representative pictures from B) LL+BL (score = 3) and C) TT+BT (score = 1) individuals are depicted.
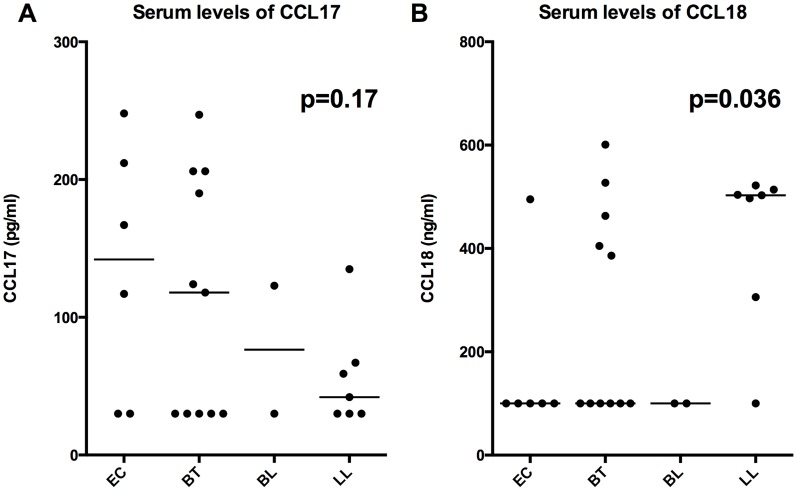
Serum CCL17 and CCL18 Protein Levels in Patients with Clinical Leprosy
Next the serum levels of CCL17 and CCL18 were compared in 6 EC (5 from Nepal and one from non-endemic area) patients without leprosy, versus 20 patients with leprosy (11 with BT, 2 with BL, and 7 with LL by RJ classification as determined by biopsy). Patients with lepromatous leprosy had a non-significant trend (p = 0.16) of decreasing CCL17 levels (Figure 5A) and a significant trend (p = 0.036) of increasing CCL18 (Figure 5B) levels in the serum.
Figure 5. CCL17 and CCL18 serum levels.
Serum levels of CCL17 and CCL18 protein were measured in leprosy patients with lesions defined by RJ classification (n = 20) compared to EC (n = 6). P values by non-parametric trend test are depicted. EC (patients without leprosy) were given a categorical value of 0, while those with leprosy were given increasing categorical values as progressing to LL pole. Lines depict median values.
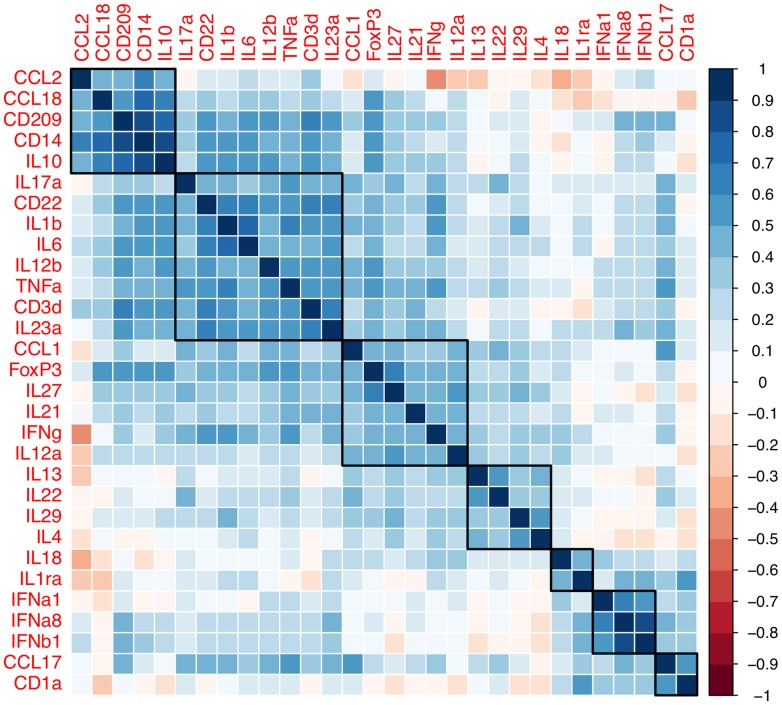
Hierarchical Clustering Determines Distinct Signatures Associated with CCL18 but Not CCL17 Expression in Dermal Biopsies
We next examined which cytokines and chemokines, correlated and clustered with cell marker levels (Figure 6). Our analyses revealed that CCL18 and IL10 expression were associated with CD209 and CD14 expression (Spearman's rho = 0.13 to 0.87), dendritic and macrophage markers respectively. In contrast, CCL17 expression was poorly correlated to all cell type markers except the dendritic and Langerhans marker, CD1a (rho = 0.53). Hierarchical clustering identified 5–7 main groups that had similar signatures (Figure 6): CCL18 group (IL10, CD209, CD14, and CCL2), TNF group (IL17a, CD22, IL1β, IL6, IL12b, CD3d, and IL23a), IFNG group (CCL1, FoxP3, IL27, IL21, and IL12a), IL4 group (IL13, IL22, and IL29 (IFNλ1)) and type I interferons (IFNA1, IFNA8, and IFNB1). CCL17 and CD1a also tended to cluster with type I interferons. These data suggest that within leprosy lesions CCL18 and CCL17 expression may be associated with specific innate immune cells rather than TH1 and TH2 T-cell cytokines traditionally thought to define leprosy (IFNG and IL4).
Figure 6. Correlation of dermal mRNA in cutaneous leprosy lesions.
Values from correlation tests (Spearman's rho statistics) are plotted from GAPDH normalized mRNA expression levels. Groups of probes were associated by hierarchical clustering using the complete-linkage clustering method (see methods).
Discussion
The primary finding of our study is that dermal mRNA levels of CCL17 and CCL18, two chemokines important in the development of a TH2 T-cell response, are associated with tuberculoid and lepromatous leprosy respectively. We also found that classical markers for leprosy and T-cell subset polarity, IL4 and IFNG, were inferior in distinguishing polar leprosy compared to CCL17 and CCL18. We also confirm an association of increased expression of IL10 within lepromatous lesions.
Our findings highlight an association of increased dermal expression of CCL17 in the skin of patients with tuberculoid leprosy. The mechanism of how CCL17 is increased in the dermal lesions of patients, who have been traditionally thought to have TH1 polar disease, is largely unknown, but there may be many possible explanations. First, increases in CCL17 expression may arise by direct stimulation of resident cells by mycobacterial antigens. Previous data suggest that CCL17 secretion from DCs is associated with increased TH1 responses [17], [18]. However, other papers suggest that CCL17 is associated with TH2 responses [13]–[15], [19]. Direct stimulation of CCL17 mRNA expression by mycobacterial antigens would suggest that the response to M. leprae in patients with TT and BT disease may have more of a TH2 polarity than had been previously thought. To support this, there have been reports of increased CCL17 expression in dendritic cells stimulated with M. tuberculosis in mice [20], but this has not been shown in humans. Second, elevated dermal expression of CCL17 could occur following recruitment of a cell type that constitutively expresses CCL17 in patients with tuberculoid leprosy. Third, chronic M. leprae exposure could promote differentiation of resident myeloid cells into CCL17-secreting cells that are more apt to control M. leprae replication. These second and third mechanisms would require there being significant differences in innate M. leprae detection between individuals with tuberculoid and lepromatous leprosy leading to either differences in cellular differentiation, or differences in the release of soluble mediators by resident cells that would influence differentiation or recruitment of new cells to lesions. Recent data suggest that NOD2 regulates M. leprae induced differentiation of myeloid precursors into cells associated with tuberculoid lesions [29]. Whether the immune cell associated with tuberculoid lesions (thought to be CD1b(+)) more actively produce CCL17 is currently unknown. Ultimately the mechanism that underlies the association of increased expression of CCL17 in tuberculoid lesions will need to be elucidated in future research.
The mechanism behind the association of increased CCL18 expression in lepromatous lesions is similarly unclear. Our clustering data suggest that CCL18 expression is associated with the expression patterns of IL10, CD209 and CD14 in lepromatous lesions, possibly implying that recruitment or differentiation of a specific cell type may be one mechanism to support the increased expression of CCL18. In support of this hypothesis, studies have described CD14+ and CD209+ monocyte-derived skin antigen presenting cells that have an immature or tolerant phenotype in normal human skin [30], [31] and have been associated with the production of CCL18 [32]. Whether these cells exist in lepromatous leprosy lesions, is not known. Mycobacterial antigens present in leprosy lesions could also stimulate the direct production of CCL18, since this has been described in monocyte derived macrophages and primary alveolar macrophages [22]. Furthermore the release of CCL18 in lepromatous lesions may lead to the development of more tolerant cells through a positive feedback loop [23], by promoting the differentiation of myeloid suppressor cells. These myeloid suppressor cells are thought to suppress the protective immune response in human T cells malignancies, and one could speculate that these cells could have a similar role in lepromatous leprosy. Furthermore, evidence that CCL18 may play a direct role in leprosy is suggested by linkage and genome wide association studies that show several CCL18 polymorphisms are associated with development of leprosy [33], [34]. In the GWAS study, three of eight CCL18 polymorphisms were significantly associated with susceptibility to leprosy using a conventional significance threshold (but not GWAS level significance) [34]. Recent data suggest that CCL18 is a ligand for CCR8 and that it induces chemotaxis of TH2 polarized T cells [35]. Whether or not CCL18 has primary effects on lepromatous leprosy development and persistence is currently unknown. However, these data suggest that CCL18 may propagate lepromatous leprosy by recruiting TH2 T cells.
Our data show trends in levels of serum CCL18 and CCL17 that match clinical findings observed with mRNA levels in skin lesions and immunohistochemical analysis (for CCL18). Our study may be underpowered to determine a significant association between CCL18 and CCL17 serum levels and leprosy phenotype. Confirmation in larger studies will be needed to determine whether there is a correlation with increasing CCL18 and declining CCL17 levels in patients with lepromatous disease.
The association of CCL17 and CCL18 expression with younger individuals is an interesting result. While these data may suggest an influence of age on the dermal expression of these two cytokine, these data may also suggest an influence of the BCG vaccine that was uniformly administered by a nationwide program that began in Nepal in 1966. Our study did not record the vaccine status of the tested individuals. Further studies will be needed to determine whether expression of CCL17 and CCL18 is modulated by age or BCG vaccination.
There are several limitations to our study. First, a potential limitation is biopsy sampling error. We isolated RNA from biopsies that were adjacent to areas used to histologically classify the patient, and these two biopsies may have been significantly different. Although we cannot exclude that possibility, this limitation would apply to any leprosy biopsy study. Our patients were diagnosed comprehensively based upon slit skin smear, skin biopsy histopathology, clinical exam and neuropathy assessments; so there were unlikely to be large misclassifications due to biopsy sampling errors. Another possible limitation of this study is type II (false positive) error due to population bias. The population studied was recruited at a tertiary care center for leprosy, and contained a large percentage of individuals who were simultaneously undergoing Type I reversal and Type II ENL reactions. This bias may influence the types of cytokines and chemokines that are significantly different. Our analysis, however, showed that the main differences (CCL17, CCL18, and CD14) were preserved despite the presence or absence of reactive states in individuals.
Interestingly, our study demonstrated that CCL17 and CCL18 distinguished leprosy polarity with greater accuracy than the traditional TH1 and TH2 cytokines (IL10, IFNG). Although this difference may be due to a stronger biologic association of CCL17 and CCL18 with polarity, other explanations are also possible. First, many previous studies had small sample sizes and compared patients at the extreme end of the leprosy poles (for example, LL versus BT and TT) and did not include as many patients with the entire spectrum of leprosy as our study did. In addition, our samples did not proportionately represent the leprosy poles with many more LL (n = 27 or 61% of the lepromatous pole patients) than TT patients (n = 3, or 8% of the tuberculoid pole patients). These proportions could potentially skew the data in the favor of LL associations and weaken TT associations of traditional cytokines (IFNG with TH1 and tuberculoid leprosy).
Supporting Information
Correlation between Fluidigm gene chip analysis and Singleplex Real-Time PCR analysis. Single assay probes for CCL17 (A) and CCL18 (B) that were normalized to GAPDH expression using standard RT-PCRa Fluidigmmicrofluidic platform. R2 values represent standard linear correlation.
(TIFF)
Primer-Probe sets used in this study. A list of sequences of the primer and probe sets used in the paper. Probes were modified by 6-carboxyfluorescein (56-FAM) on the 5′ end and Iowa Black Quencher (3IABkFQ) on the 3′ end. The dyes also contain an additional internal quencher (ZEN).
(DOCX)
Acknowledgments
We thank the patients and staff at Anandaban Hospital for the clinical work associated with this study. We thank Saraswoti Khadge and Pratibha Thapa for technical assistance. We also thank Dr. Lakshmi Rajan and Dr. Joyce Ponnaiya of the Schieffelin Institute of Health – Research & Leprosy Centre, Karigiri, Tamil Nadu, India for their expert histopathological services. We thank Kelly Hudkins and Charles Alpers for assistance with CCL17 and CCL18 immunohistochemistry.
Funding Statement
Funding for this study was due to the contributions of the Heiser Foundation. This work was also supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health K08 [AI080952 for WRB, and 1K24AI089794-for TRH], The Heiser Program for Research in Tuberculosis and Leprosy with a grant to WRB, the Burroughs Wellcome Fund to TRH, and The Leprosy Mission International to DAH and Q.M. Gastmann-Wichers Foundation, Maltalep and the Dutch Leprosy Relief association (NLR) for AG and SJFvdE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, et al. (2006) The continuing challenges of leprosy. Clinical Microbiology Reviews 19: 338–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Misch EA, Berrington WR, Vary JC Jr, Hawn TR (2010) Leprosy and the human genome. Microbiology and molecular biology reviews: MMBR 74: 589–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ridley DS, Jopling WH (1966) Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis 34: 255–273. [PubMed] [Google Scholar]
- 4. Yamamura M, Uyemura K, Deans RJ, Weinberg K, Rea TH, et al. (1991) Defining Protective Responses to Pathogens - Cytokine Profiles in Leprosy Lesions. Science 254: 277–279. [DOI] [PubMed] [Google Scholar]
- 5. Van Voorhis WC, Kaplan G, Sarno EN, Horwitz MA, Steinman RM, et al. (1982) The cutaneous infiltrates of leprosy: cellular characteristics and the predominant T-cell phenotypes. N Engl J Med 307: 1593–1597. [DOI] [PubMed] [Google Scholar]
- 6. Bleharski JR, Li H, Meinken C, Graeber TG, Ochoa MT, et al. (2003) Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science 301: 1527–1530. [DOI] [PubMed] [Google Scholar]
- 7. Modlin RL (2010) The innate immune response in leprosy. Curr Opin Immunol 22: 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ottenhoff TH (2012) New pathways of protective and pathological host defense to mycobacteria. Trends Microbiol 20: 419–428. [DOI] [PubMed] [Google Scholar]
- 9. de Nadai P, Charbonnier AS, Chenivesse C, Senechal S, Fournier C, et al. (2006) Involvement of CCL18 in allergic asthma. Journal of immunology 176: 6286–6293. [DOI] [PubMed] [Google Scholar]
- 10. Gunther C, Bello-Fernandez C, Kopp T, Kund J, Carballido-Perrig N, et al. (2005) CCL18 is expressed in atopic dermatitis and mediates skin homing of human memory T cells. Journal of immunology 174: 1723–1728. [DOI] [PubMed] [Google Scholar]
- 11. Kim HB, Kim CK, Iijima K, Kobayashi T, Kita H (2009) Protein microarray analysis in patients with asthma: elevation of the chemokine PARC/CCL18 in sputum. Chest 135: 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prasse A, Pechkovsky DV, Toews GB, Schafer M, Eggeling S, et al. (2007) CCL18 as an indicator of pulmonary fibrotic activity in idiopathic interstitial pneumonias and systemic sclerosis. Arthritis Rheum 56: 1685–1693. [DOI] [PubMed] [Google Scholar]
- 13. Belperio JA, Dy M, Murray L, Burdick MD, Xue YY, et al. (2004) The role of the Th2 CC chemokine ligand CCL17 in pulmonary fibrosis. Journal of immunology 173: 4692–4698. [DOI] [PubMed] [Google Scholar]
- 14. Fujisawa T, Nagao M, Hiraguchi Y, Katsumata H, Nishimori H, et al. (2009) Serum measurement of thymus and activation-regulated chemokine/CCL17 in children with atopic dermatitis: elevated normal levels in infancy and age-specific analysis in atopic dermatitis. Pediatr Allergy Immunol 20: 633–641. [DOI] [PubMed] [Google Scholar]
- 15. Riis JL, Johansen C, Vestergaard C, Bech R, Kragballe K, et al. (2011) Kinetics and differential expression of the skin-related chemokines CCL27 and CCL17 in psoriasis, atopic dermatitis and allergic contact dermatitis. Experimental dermatology 20: 789–794. [DOI] [PubMed] [Google Scholar]
- 16. Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, et al. (2009) Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc Natl Acad Sci U S A 106: 14978–14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alferink J, Lieberam I, Reindl W, Behrens A, Weiss S, et al. (2003) Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. J Exp Med 197: 585–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henry CJ, Ornelles DA, Mitchell LM, Brzoza-Lewis KL, Hiltbold EM (2008) IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J Immunol 181: 8576–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawasaki S, Takizawa H, Yoneyama H, Nakayama T, Fujisawa R, et al. (2001) Intervention of thymus and activation-regulated chemokine attenuates the development of allergic airway inflammation and hyperresponsiveness in mice. J Immunol 166: 2055–2062. [DOI] [PubMed] [Google Scholar]
- 20. Jang S, Uzelac A, Salgame P (2008) Distinct chemokine and cytokine gene expression pattern of murine dendritic cells and macrophages in response to Mycobacterium tuberculosis infection. J Leukoc Biol 84: 1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park CO, Lee HJ, Lee JH, Wu WH, Chang NS, et al. (2008) Increased expression of CC chemokine ligand 18 in extrinsic atopic dermatitis patients. Experimental dermatology 17: 24–29. [DOI] [PubMed] [Google Scholar]
- 22. Ferrara G, Bleck B, Richeldi L, Reibman J, Fabbri LM, et al. (2008) Mycobacterium tuberculosis induces CCL18 expression in human macrophages. Scand J Immunol 68: 668–674. [DOI] [PubMed] [Google Scholar]
- 23. Schraufstatter IU, Zhao M, Khaldoyanidi SK, Discipio RG (2012) The chemokine CCL18 causes maturation of cultured monocytes to macrophages in the M2 spectrum. Immunology 135: 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jha PK, Gurung R, Gyawali N, Nepal HP, Baral DD, et al. (2012) Tuberculin reactivity in bacille calmette-guerin vaccinated individuals with sputum positive pulmonary tuberculosis. Journal of College of Medical Sciences-Nepal 7. [Google Scholar]
- 25.Hartigan JA (1975) Clustering algorithms. New York,: Wiley. xiii, 351 p. p. [Google Scholar]
- 26. McQuitty LL (1960) Hierarchical Linkage Analysis for the Isolation of Types. Educational and Psychological Measurement 20: 55–67. [Google Scholar]
- 27. Friendly M (2002) Corrgrams: Exploratory displays for correlation matrices. The American Statistician 56: 316–324. [Google Scholar]
- 28. Murdoch DJ, Chow ED (1996) A graphical display of large correlation matrices. The American Statistician 50: 178–180. [Google Scholar]
- 29. Schenk M, Krutzik SR, Sieling PA, Lee DJ, Teles RM, et al. (2012) NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat Med 18: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ochoa MT, Loncaric A, Krutzik SR, Becker TC, Modlin RL (2008) “Dermal dendritic cells” comprise two distinct populations: CD1+ dendritic cells and CD209+ macrophages. The Journal of investigative dermatology 128: 2225–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Angel CE, Lala A, Chen CJ, Edgar SG, Ostrovsky LL, et al. (2007) CD14+ antigen-presenting cells in human dermis are less mature than their CD1a+ counterparts. Int Immunol 19: 1271–1279. [DOI] [PubMed] [Google Scholar]
- 32. Gustafsson C, Mjosberg J, Matussek A, Geffers R, Matthiesen L, et al. (2008) Gene expression profiling of human decidual macrophages: evidence for immunosuppressive phenotype. PLoS One 3: e2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jamieson SE, Miller EN, Black GF, Peacock CS, Cordell HJ, et al. (2004) Evidence for a cluster of genes on chromosome 17q11-q21 controlling susceptibility to tuberculosis and leprosy in Brazilians. Genes and immunity 5: 46–57. [DOI] [PubMed] [Google Scholar]
- 34. Zhang FR, Huang W, Chen SM, Sun LD, Liu H, et al. (2009) Genomewide association study of leprosy. N Engl J Med 361: 2609–2618. [DOI] [PubMed] [Google Scholar]
- 35. Islam SA, Ling MF, Leung J, Shreffler WG, Luster AD (2013) Identification of human CCR8 as a CCL18 receptor. J Exp Med [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between Fluidigm gene chip analysis and Singleplex Real-Time PCR analysis. Single assay probes for CCL17 (A) and CCL18 (B) that were normalized to GAPDH expression using standard RT-PCRa Fluidigmmicrofluidic platform. R2 values represent standard linear correlation.
(TIFF)
Primer-Probe sets used in this study. A list of sequences of the primer and probe sets used in the paper. Probes were modified by 6-carboxyfluorescein (56-FAM) on the 5′ end and Iowa Black Quencher (3IABkFQ) on the 3′ end. The dyes also contain an additional internal quencher (ZEN).
(DOCX)